www.cell.com/cell/fulltex...
Key findings in a thread (1/6):

www.cell.com/cell/fulltex...
Key findings in a thread (1/6):
He joins EMBL from @mpi-cbg.de in Dresden. He is also Professor of Molecular Biology @tudresden.bsky.social, and was a group leader at EMBL Heidelberg from 1993 to 1999.
www.embl.org/news/people-...

He joins EMBL from @mpi-cbg.de in Dresden. He is also Professor of Molecular Biology @tudresden.bsky.social, and was a group leader at EMBL Heidelberg from 1993 to 1999.
www.embl.org/news/people-...
✅ Free
🔓 Open Source
🧬 More cloning methods than SnapGene
🤖 Can be automated with python
👨🔬 Built by a researcher — for researchers!
👉 Check it out at opencloning.org

✅ Free
🔓 Open Source
🧬 More cloning methods than SnapGene
🤖 Can be automated with python
👨🔬 Built by a researcher — for researchers!
👉 Check it out at opencloning.org
1) active histone mods occur independently of transcription
2) transcription coordinates histone deacetylation at active promoters
www.science.org/doi/10.1126/...

1) active histone mods occur independently of transcription
2) transcription coordinates histone deacetylation at active promoters
www.science.org/doi/10.1126/...

Fully funded postdoc position in my group!
humantechnopole.it/en/research-...
Fully funded postdoc position in my group!
humantechnopole.it/en/research-...


What then makes low-affinity motifs important for enhancer regulation?

What then makes low-affinity motifs important for enhancer regulation?
The cis-regulatory logic integrating spatial and temporal patterning in the vertebrate neural tube
A global temporal chromatin program operates across the vertebrate nervous system to control neural cell diversity
www.cell.com/developmenta...

The cis-regulatory logic integrating spatial and temporal patterning in the vertebrate neural tube
A global temporal chromatin program operates across the vertebrate nervous system to control neural cell diversity
www.cell.com/developmenta...

cheers to the @erin-schuman.bsky.social & @debbysilver.bsky.social labs!
www.nature.com/articles/s41...
cheers to the @erin-schuman.bsky.social & @debbysilver.bsky.social labs!
www.nature.com/articles/s41...
(TLDR; low-affinity motifs matter as pioneers!)

(TLDR; low-affinity motifs matter as pioneers!)


Here is how and why I gave this model organism a visual upgrade 🧵(1/7)

Here is how and why I gave this model organism a visual upgrade 🧵(1/7)
@ScienceAdvances
where we dissect the gene regulatory landscape guiding mouse sex determination!
science.org/doi/10.1126/...
A huge well done to @IsabelleStevant.genomic.social.ap.brid.gy, Meshi and Elisheva who spearhead this project!
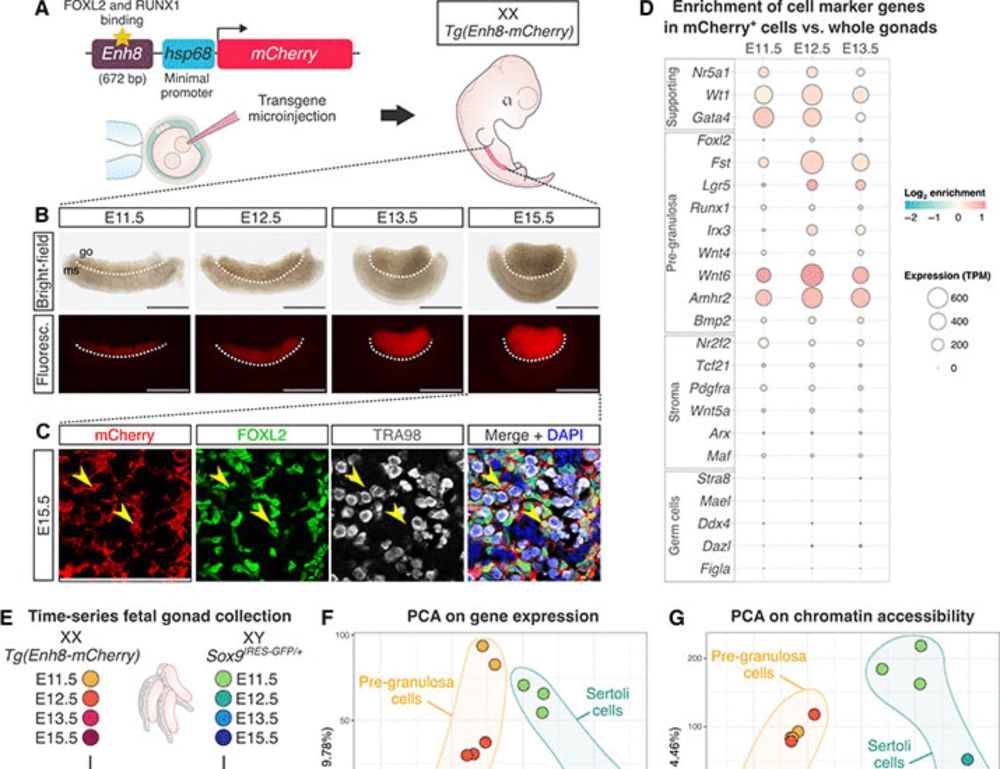
@ScienceAdvances
where we dissect the gene regulatory landscape guiding mouse sex determination!
science.org/doi/10.1126/...
A huge well done to @IsabelleStevant.genomic.social.ap.brid.gy, Meshi and Elisheva who spearhead this project!
www.nature.com/articles/s41...

www.nature.com/articles/s41...
It was a long, 7 years long journey, which coordinated efforts of 50+ researchers, proud to be on of them.
More results from Codebook about poorly studied TFs are coming soon.
It was a long, 7 years long journey, which coordinated efforts of 50+ researchers, proud to be on of them.
More results from Codebook about poorly studied TFs are coming soon.
Next X-inactivation meeting in Sapporo, Japan, 19-23 October 2026. Visit x-inactivation-meeting.org to join our mailing list. 🧬 speakers @dandergassen.bsky.social @marnieblewitt.bsky.social @heard65.bsky.social @crougeulle.bsky.social @sexchrlab.bsky.social @zhouqi1982.bsky.social

Next X-inactivation meeting in Sapporo, Japan, 19-23 October 2026. Visit x-inactivation-meeting.org to join our mailing list. 🧬 speakers @dandergassen.bsky.social @marnieblewitt.bsky.social @heard65.bsky.social @crougeulle.bsky.social @sexchrlab.bsky.social @zhouqi1982.bsky.social
openrxiv.org/openrxiv-day/
openrxiv.org/openrxiv-day/
It turns out you can integrate arrays with super high efficiency using PhiC31.
www.biorxiv.org/content/10.1...

It turns out you can integrate arrays with super high efficiency using PhiC31.
www.biorxiv.org/content/10.1...


