
"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)

"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
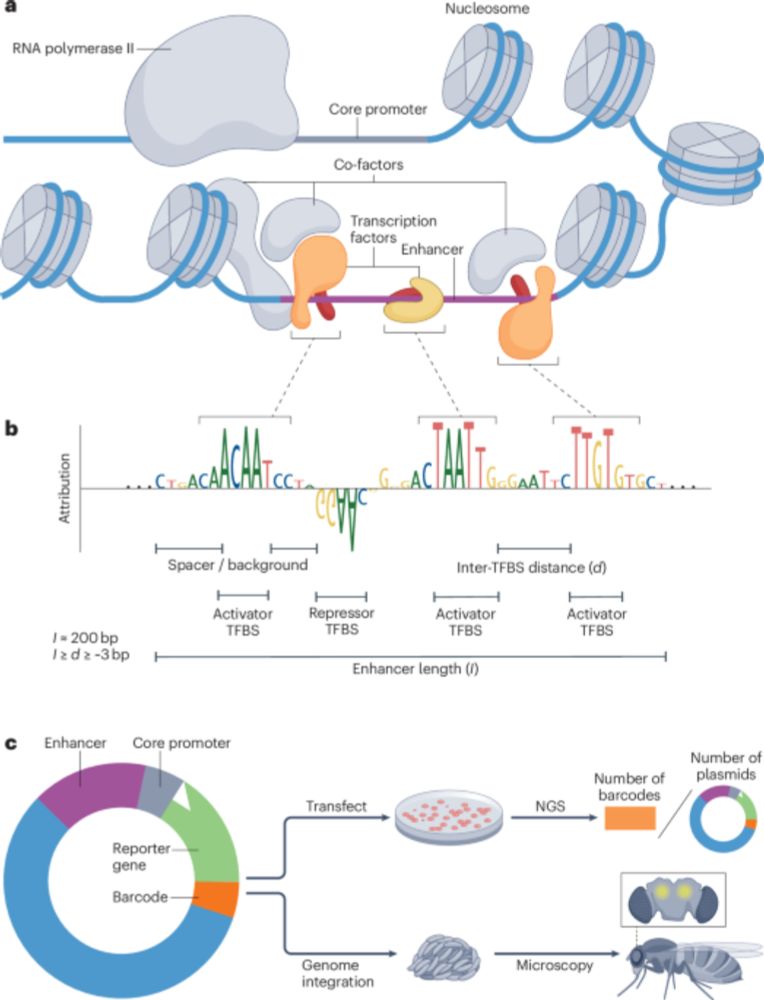
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
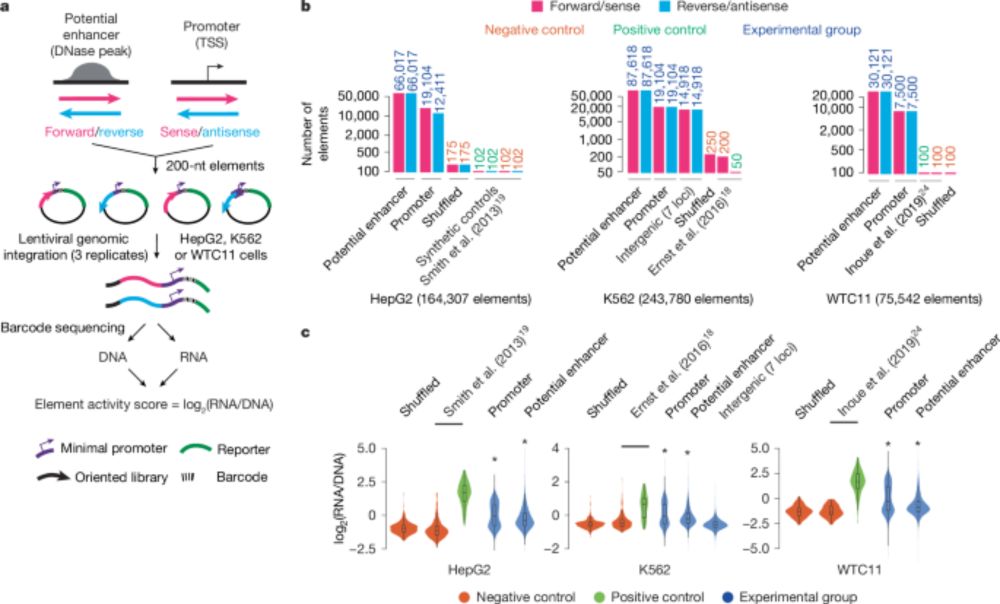
www.biorxiv.org/content/10.1...
A 🧵about what it can do:
#SynBio #DeepLearning #GeneRegulation

www.biorxiv.org/content/10.1...
A 🧵about what it can do:
#SynBio #DeepLearning #GeneRegulation
Just two weeks ago I was explaining to a junior colleague the problem of exaggerated claims in science. This paragraph is exactly what should be printed in place of a user agreement when anybody submits a paper.
it's so true and hits so hard:
Just two weeks ago I was explaining to a junior colleague the problem of exaggerated claims in science. This paragraph is exactly what should be printed in place of a user agreement when anybody submits a paper.
Join us for our next Kipoi Seminar with with Dmitry Penzar,
@pensarata.bsky.social @ autosome.org!
👉LegNet: parameter-efficient modeling of gene regulatory regions using modern convolutional neural network
📅Wed Dec 4, 5:30pm CET
🧬 kipoi.org/seminar/
Join us for our next Kipoi Seminar with with Dmitry Penzar,
@pensarata.bsky.social @ autosome.org!
👉LegNet: parameter-efficient modeling of gene regulatory regions using modern convolutional neural network
📅Wed Dec 4, 5:30pm CET
🧬 kipoi.org/seminar/

biorxiv.org/cgi/content/...
Read more below!
1/12

biorxiv.org/cgi/content/...
Read more below!
1/12
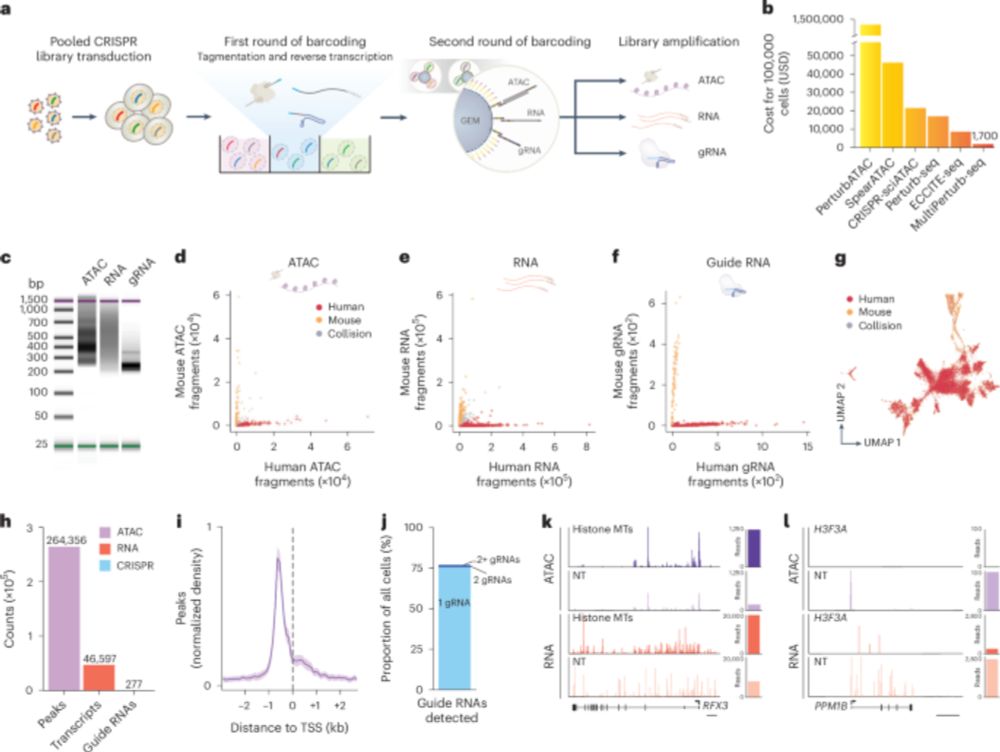
arxiv.org/abs/2411.11158

Paper here: www.biorxiv.org/content/10.1...
1/
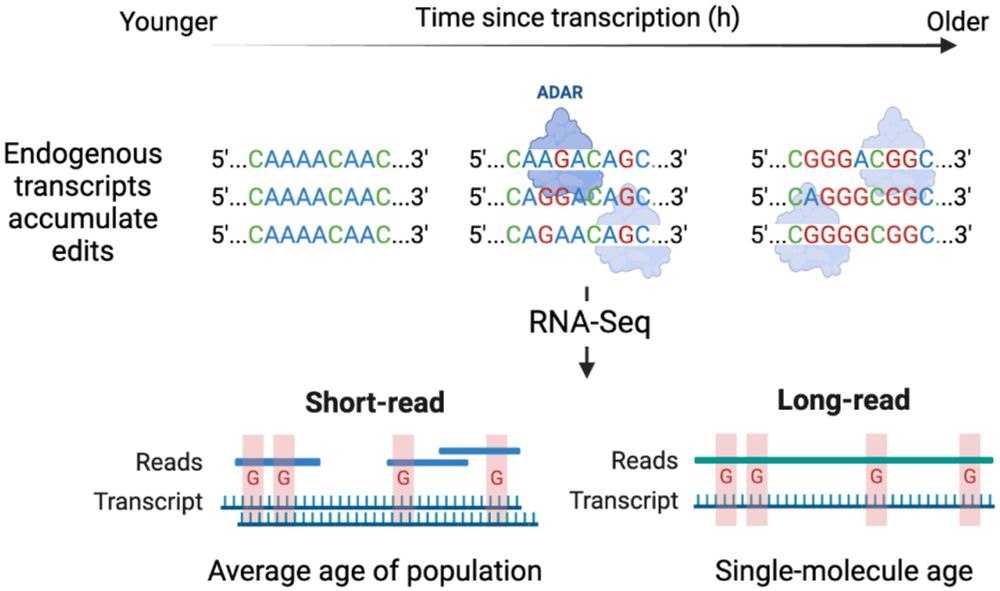
Paper here: www.biorxiv.org/content/10.1...
1/





www.nature.com/articles/s41...

www.nature.com/articles/s41...


