
It's the first tool that builds on Sassy, the approximate-DNA-searching tool that @rickbitloo.bsky.social and myself developed earlier this year, specifically with this application in mind.
www.biorxiv.org/content/10.1...
Want to get started? github.com/rickbeeloo/b...
It's the first tool that builds on Sassy, the approximate-DNA-searching tool that @rickbitloo.bsky.social and myself developed earlier this year, specifically with this application in mind.

Aardvark: Sifting through differences in a mound of variants
GitHub: github.com/PacificBiosc...
Some highlights in this thread:
1/N
Aardvark: Sifting through differences in a mound of variants
GitHub: github.com/PacificBiosc...
Some highlights in this thread:
1/N
genomebiology.biomedcentral.com/articles/10....
genomebiology.biomedcentral.com/articles/10....
Mapping long reads to pangenome graphs is ~10x faster than with GraphAligner, with veeery slightly better mapping accuracy, short variant calling, and SV genotyping than GraphAligner or Minimap2
Mapping long reads to pangenome graphs is ~10x faster than with GraphAligner, with veeery slightly better mapping accuracy, short variant calling, and SV genotyping than GraphAligner or Minimap2
Until now, using AccuSNV.
Herui Liao trained an ML model based on our previous meticulously called SNVs.
www.biorxiv.org/content/10.1...

Until now, using AccuSNV.
Herui Liao trained an ML model based on our previous meticulously called SNVs.
www.biorxiv.org/content/10.1...
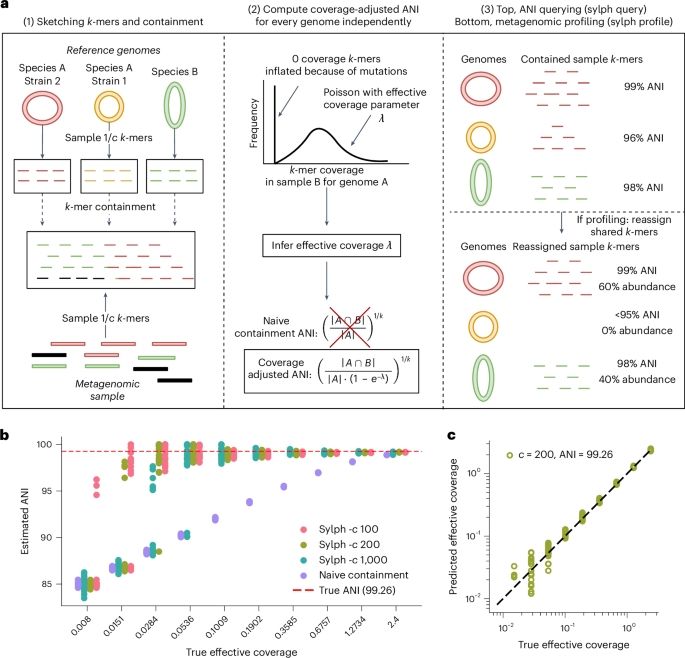
Computing average nucleotide identity (ANI) is neither conceptually nor computationally trivial. Its definition has evolved over years, with different meanings and assumptions (1/5)

Computing average nucleotide identity (ANI) is neither conceptually nor computationally trivial. Its definition has evolved over years, with different meanings and assumptions (1/5)
www.biorxiv.org/content/10.1...
#Bioinformatics #genomics #assembly #assemblygraphs #software

www.biorxiv.org/content/10.1...
#Bioinformatics #genomics #assembly #assemblygraphs #software
lh3.github.io/2025/09/11/a...

lh3.github.io/2025/09/11/a...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...

It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
nature.com/articles/s44...

nature.com/articles/s44...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
rdcu.be/eCbj4
rdcu.be/eCbj4
Read all about it here: nature.com/articles/s41...
Great collab with Chew and Hirao lab
x.com/NiranjanTW/s...

Read all about it here: nature.com/articles/s41...
Great collab with Chew and Hirao lab
x.com/NiranjanTW/s...
Github: github.com/vikshiv/mume...
First paper: genomebiology.biomedcentral.com/articles/10....
Second: www.biorxiv.org/content/10.1... #WABI2025



Github: github.com/vikshiv/mume...
First paper: genomebiology.biomedcentral.com/articles/10....
Second: www.biorxiv.org/content/10.1... #WABI2025
Proud to have contributed to Metalog, the latest @borklab.bsky.social resource:
www.biorxiv.org/content/10.1...
#microsky #microbiome
Proud to have contributed to Metalog, the latest @borklab.bsky.social resource:
www.biorxiv.org/content/10.1...
#microsky #microbiome
and GH repo: github.com/refresh-bio/...

and GH repo: github.com/refresh-bio/...

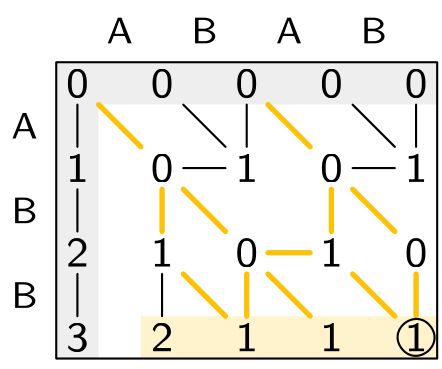
Ever need to search for approximate matches of short DNA strings?
Sassy is the tool to use!
Available now wherever you get your code
With @rickbitloo.bsky.social
curiouscoding.nl/papers/sassy...
github.com/ragnarGrootK...
Ever need to search for approximate matches of short DNA strings?
Sassy is the tool to use!
Available now wherever you get your code
With @rickbitloo.bsky.social
curiouscoding.nl/papers/sassy...
github.com/ragnarGrootK...
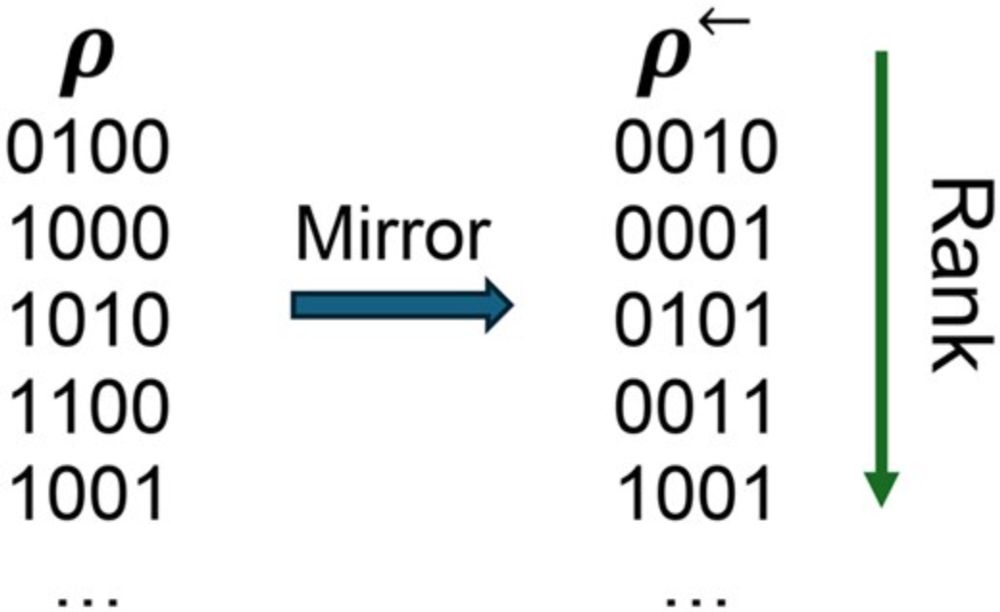
25 Additional Problems -- Extension to the Book "125 Problems in Text Algorithms"
https://arxiv.org/abs/2507.05770
25 Additional Problems -- Extension to the Book "125 Problems in Text Algorithms"
https://arxiv.org/abs/2507.05770


