Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Please try, we need feedback 🧪🧶🧬🔬

Please try, we need feedback 🧪🧶🧬🔬


Read more: www2.mrc-lmb.cam.ac.uk/protein-sort...
Read more: www2.mrc-lmb.cam.ac.uk/protein-sort...
buff.ly/j3TSIkn
Happy (but mostly relieved) to share my dream project with @boudkerlab.bsky.social, now published in @natsmb.nature.com! We used evolution, protein engineering & cryoEM to uncover how ion coupling in glutamate transporters works, and how it evolved.🧵
Free article: go.nature.com/4oRUC1q

Happy (but mostly relieved) to share my dream project with @boudkerlab.bsky.social, now published in @natsmb.nature.com! We used evolution, protein engineering & cryoEM to uncover how ion coupling in glutamate transporters works, and how it evolved.🧵
Free article: go.nature.com/4oRUC1q
www.cell.com/cell-reports...
#cryoEM @dshatskiy.bsky.social @jakecolautti.bsky.social

www.cell.com/cell-reports...
#cryoEM @dshatskiy.bsky.social @jakecolautti.bsky.social
Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social

Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social
For this call, collaborative projects between two groups will be funded. Take a look at the call and the groups and drop me an email if interested. Share with colleagues too!
www.mdc-berlin.de/postdocs#t-g...
For this call, collaborative projects between two groups will be funded. Take a look at the call and the groups and drop me an email if interested. Share with colleagues too!
www.mdc-berlin.de/postdocs#t-g...

Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM
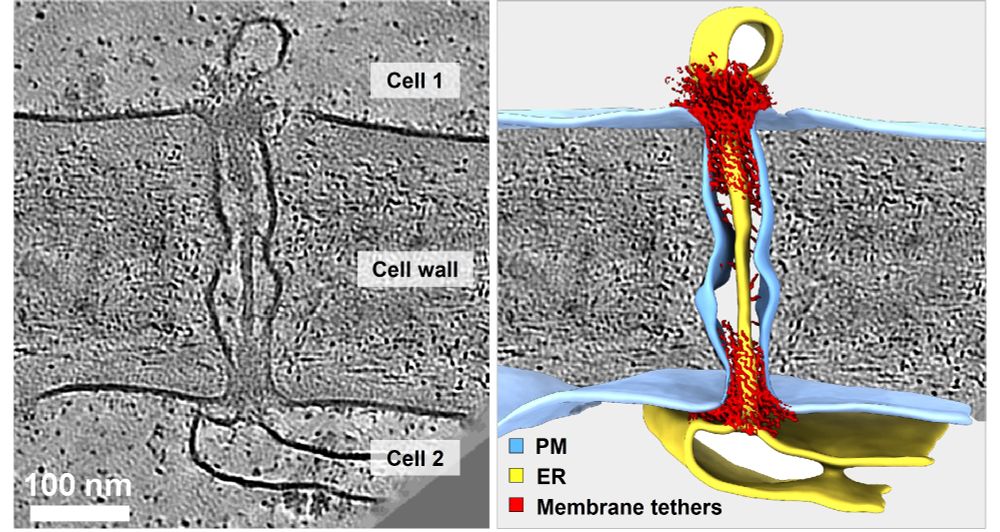
Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM

Download our code and try it yourself. 🥳
Download our code and try it yourself. 🥳
The mechanistic basis of cargo selection during Golgi maturation
Taylor, Zubkov, Ciazynska et al.
doi.org/10.1101/2025...

The mechanistic basis of cargo selection during Golgi maturation
Taylor, Zubkov, Ciazynska et al.
doi.org/10.1101/2025...
Out in @pnas.org
Congrats @janboehning.bsky.social @abultarafder.bsky.social Miles Graham @mathieucoureuil.bsky.social
@mrclmb.bsky.social @hopitalnecker.bsky.social
www.pnas.org/doi/10.1073/...
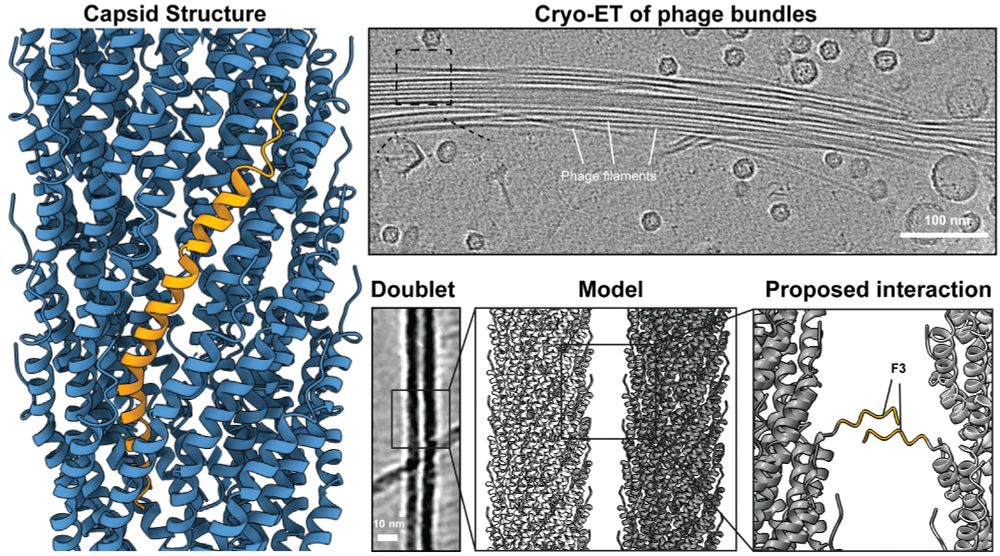
Out in @pnas.org
Congrats @janboehning.bsky.social @abultarafder.bsky.social Miles Graham @mathieucoureuil.bsky.social
@mrclmb.bsky.social @hopitalnecker.bsky.social
www.pnas.org/doi/10.1073/...

❕https://www.shawprize.org/laureates/2025-life-science-medicine/
#CryoEM #cryoET #Award

❕https://www.shawprize.org/laureates/2025-life-science-medicine/
#CryoEM #cryoET #Award
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/

Congratulations also to the #LMBAlumni joining them: John Briggs, Graham Hatfull & Baljit Khakh.
Read more: www2.mrc-lmb.cam.ac.uk/leo-james-gr...
#LMBNews

Congratulations also to the #LMBAlumni joining them: John Briggs, Graham Hatfull & Baljit Khakh.
Read more: www2.mrc-lmb.cam.ac.uk/leo-james-gr...
#LMBNews
❕https://royalsociety.org/news/2025/05/new-fellows/
@briggsgroup.bsky.social @royalsociety.org

❕https://royalsociety.org/news/2025/05/new-fellows/
@briggsgroup.bsky.social @royalsociety.org
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
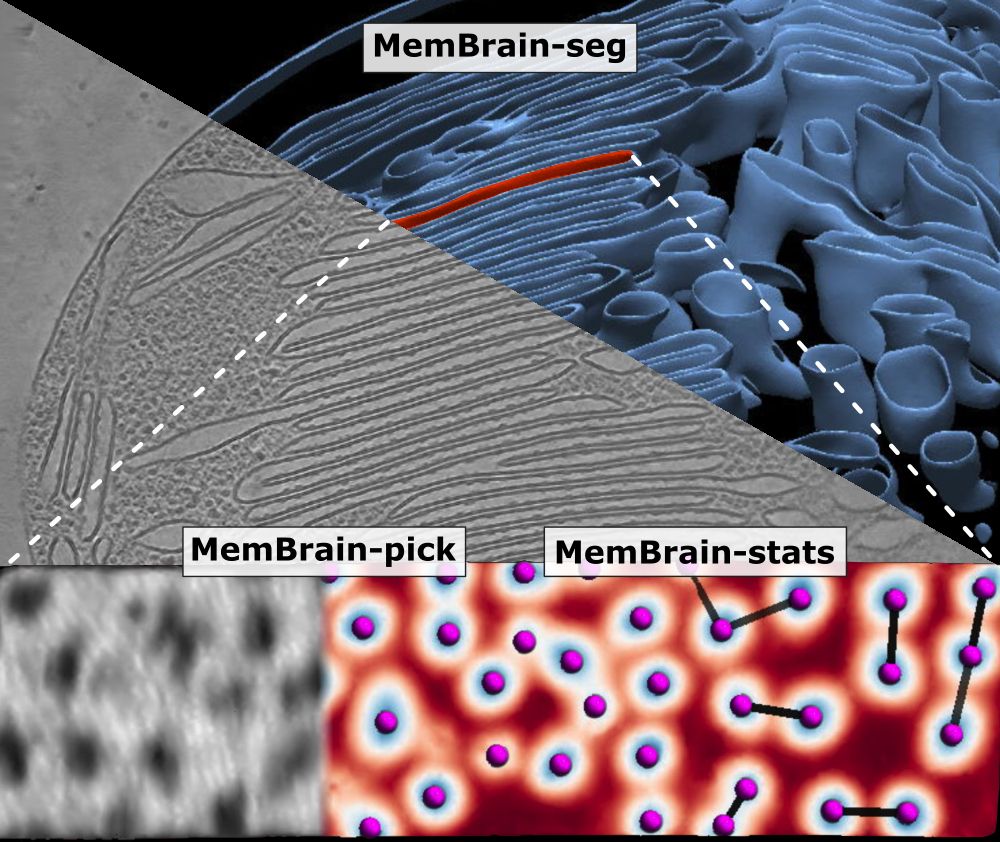
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo

