

“A Bait-and-Switch strategy links phenotypes to genes coding for Polymer-Degrading Enzymes in Intact Microbiomes.”
In this work, we discovered novel chitinases directly on microbiome; a study brilliantly led by Dr. Colleen Yancey
www.biorxiv.org/content/10.1...
“A Bait-and-Switch strategy links phenotypes to genes coding for Polymer-Degrading Enzymes in Intact Microbiomes.”
In this work, we discovered novel chitinases directly on microbiome; a study brilliantly led by Dr. Colleen Yancey
www.biorxiv.org/content/10.1...

“A Bait-and-Switch strategy links phenotypes to genes coding for Polymer-Degrading Enzymes in Intact Microbiomes.”
In this work, we discovered novel chitinases directly on microbiome; a study brilliantly led by Dr. Colleen Yancey
www.biorxiv.org/content/10.1...


Check out our latest preprint:
🧬 “A Novel NGS-Compatible Enzymatic Strategy Enables Carryover Contamination Removal and Enhances Sequencing Performance” www.biorxiv.org/content/10.1...

Check out our latest preprint:
🧬 “A Novel NGS-Compatible Enzymatic Strategy Enables Carryover Contamination Removal and Enhances Sequencing Performance” www.biorxiv.org/content/10.1...
Engineering E. coli bacteria to turn plastic waste into paracetamol (Tylenol)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
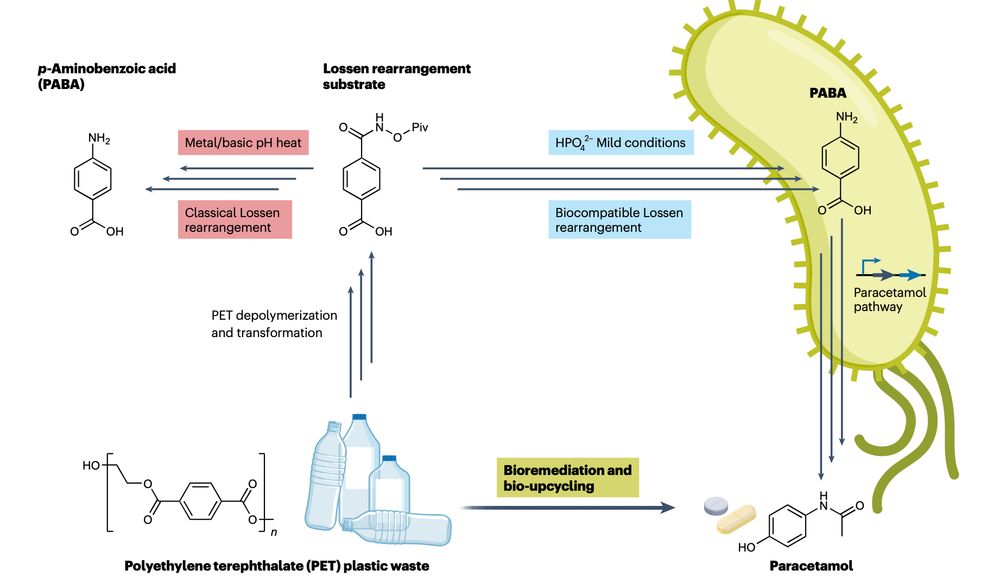
Engineering E. coli bacteria to turn plastic waste into paracetamol (Tylenol)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
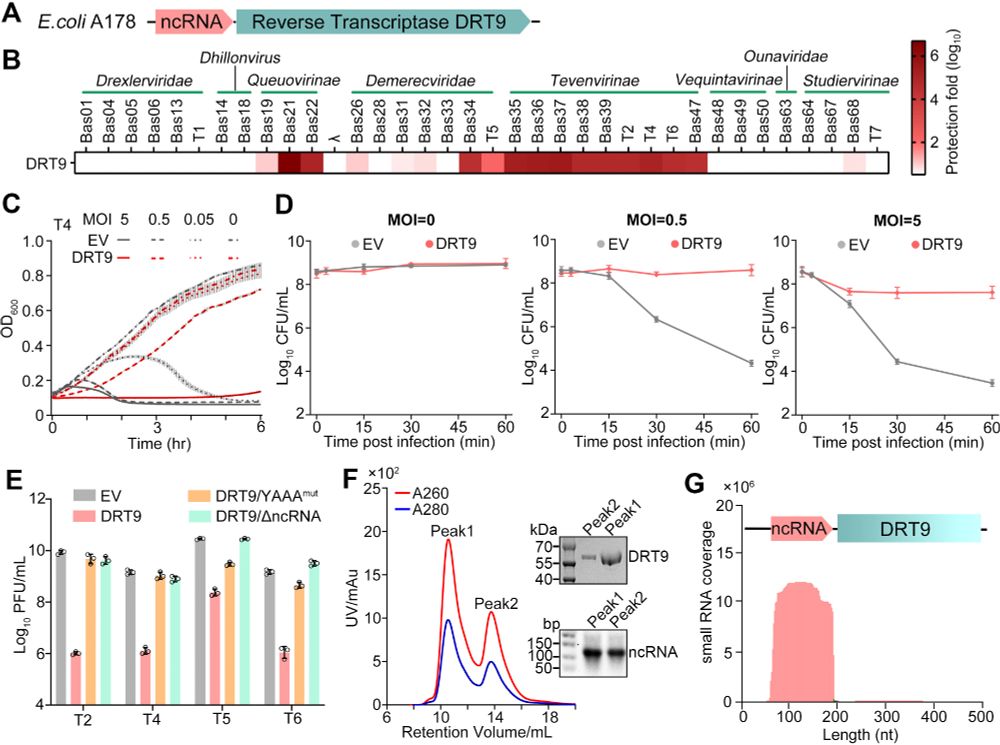
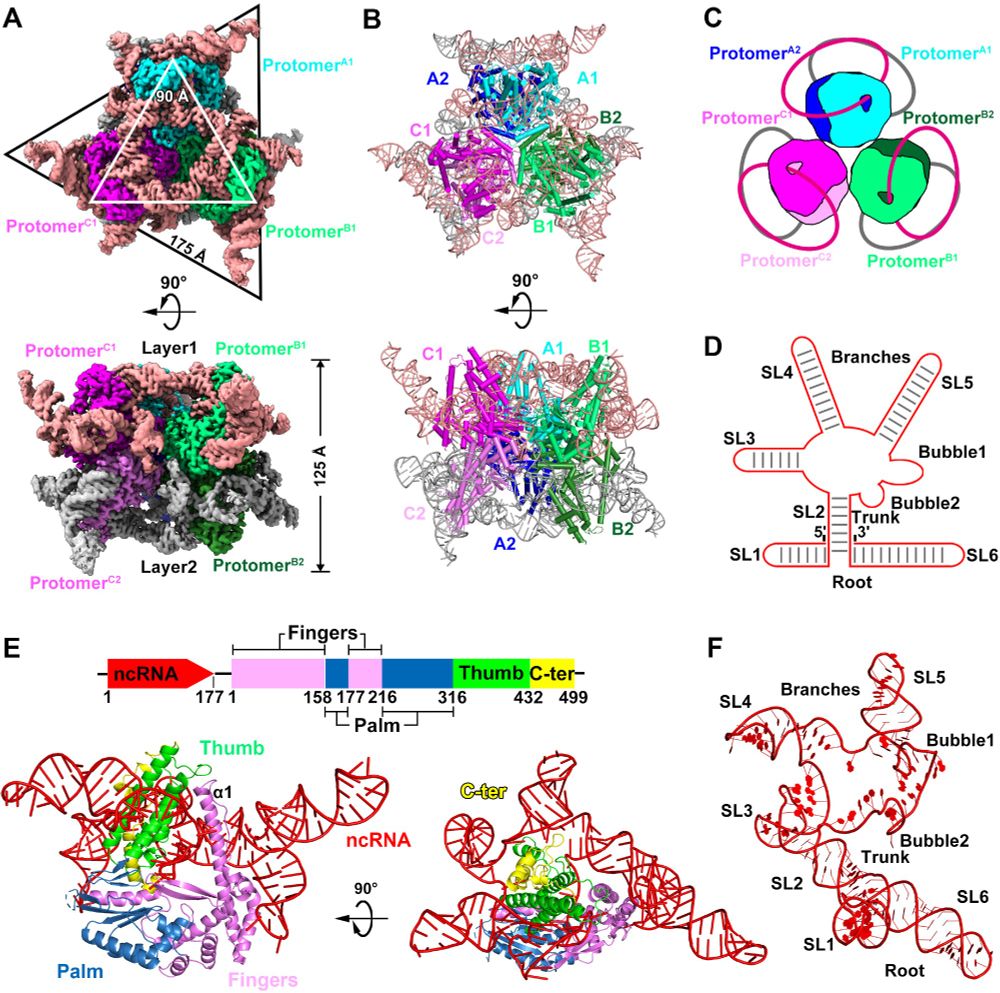
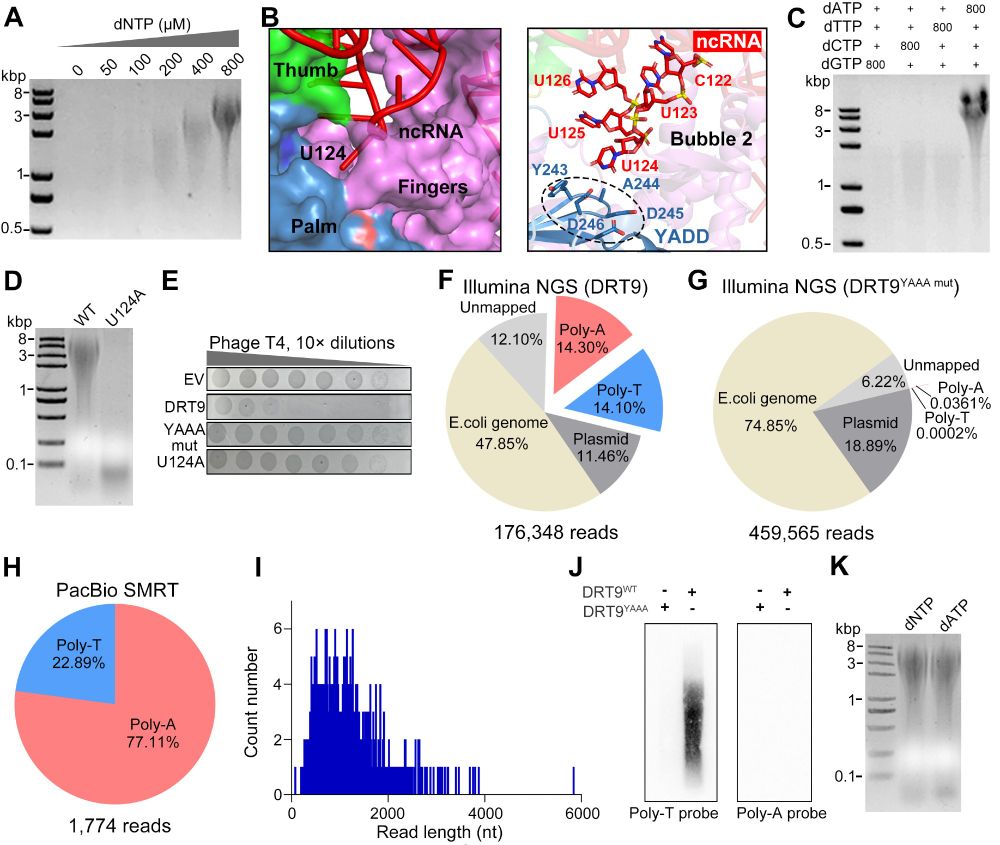
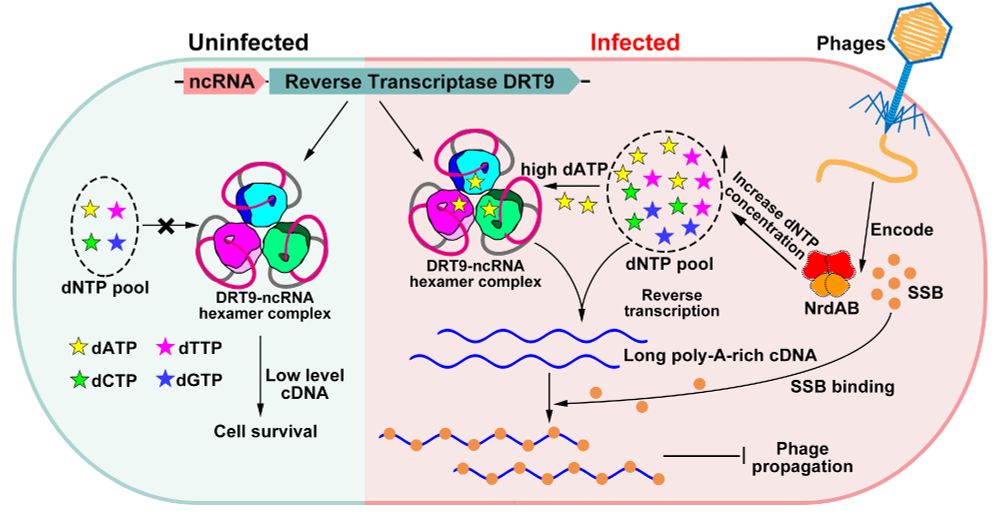
doi.org/10.1093/nar/...
doi.org/10.1093/nar/...
#Toulouse @sufs-paris-rp.bsky.social
@emmanuelleperez.bsky.social
"C'est très important d'avoir un lien très fort entre la recherche et la formation, de former à l'esprit scientifique pour se confronter à la complexité du monde d'aujourd'hui"
www.francetvinfo.fr/replay-jt/fr...

#Toulouse @sufs-paris-rp.bsky.social
@emmanuelleperez.bsky.social
"C'est très important d'avoir un lien très fort entre la recherche et la formation, de former à l'esprit scientifique pour se confronter à la complexité du monde d'aujourd'hui"
www.francetvinfo.fr/replay-jt/fr...

...BUT: they still don't capture a lot of the 🦠 species that are out there
New @jgi.doe.gov paper led by Dongying Wu and Natalia Ivanova: www.science.org/doi/10.1126/...

...BUT: they still don't capture a lot of the 🦠 species that are out there
New @jgi.doe.gov paper led by Dongying Wu and Natalia Ivanova: www.science.org/doi/10.1126/...


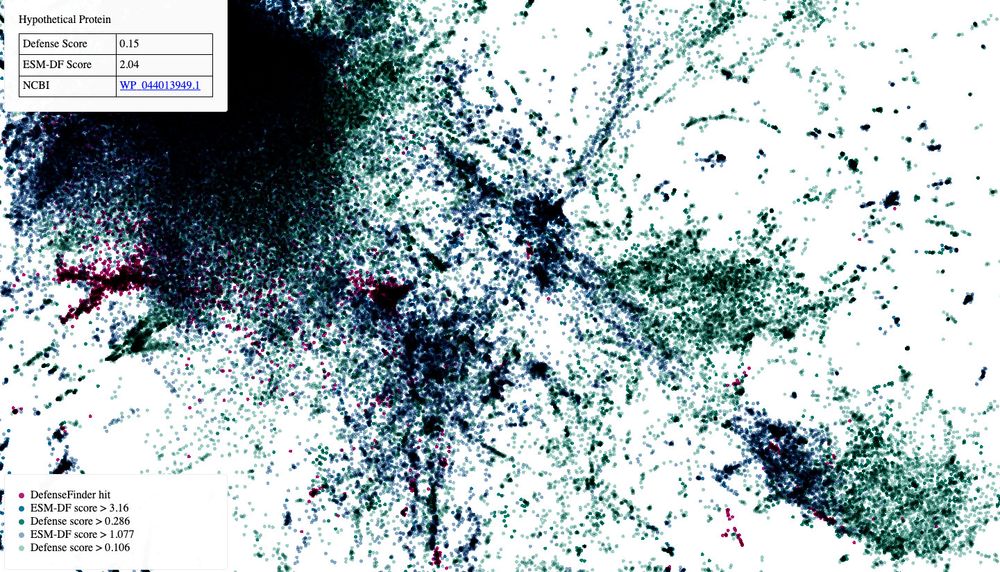
We used protein and genomic language models (and defense score!) to start bringing some answers: >45 000 protein families.
Leb by E. Mordret.
www.biorxiv.org/content/10.1...
Interactive UMAP to have fun
mdmparis.github.io/antiphage-la...
We used protein and genomic language models (and defense score!) to start bringing some answers: >45 000 protein families.
Leb by E. Mordret.
www.biorxiv.org/content/10.1...
Interactive UMAP to have fun
mdmparis.github.io/antiphage-la...
@gangfang.bsky.social @biorxivpreprint.bsky.social
t.co/QisGjqmInv
#ImmunoSky 🧪
@gangfang.bsky.social @biorxivpreprint.bsky.social
t.co/QisGjqmInv
#ImmunoSky 🧪

The #NIH has compiled over 2,000 illustrations including swatches and pattern brushes
bioart.niaid.nih.gov
#rstats #dataviz #phd #illustration

The #NIH has compiled over 2,000 illustrations including swatches and pattern brushes
bioart.niaid.nih.gov
#rstats #dataviz #phd #illustration
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
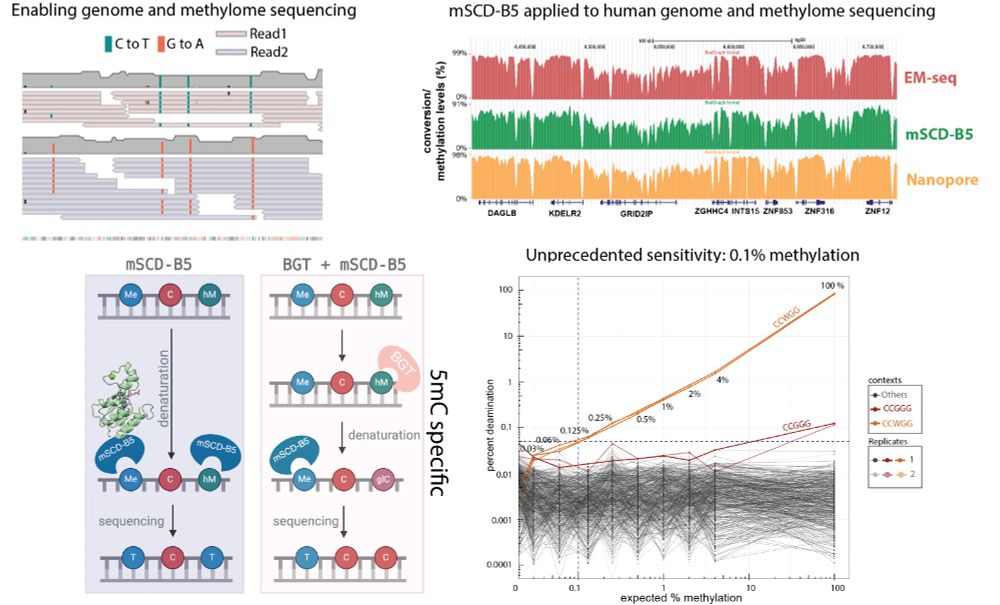
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.

