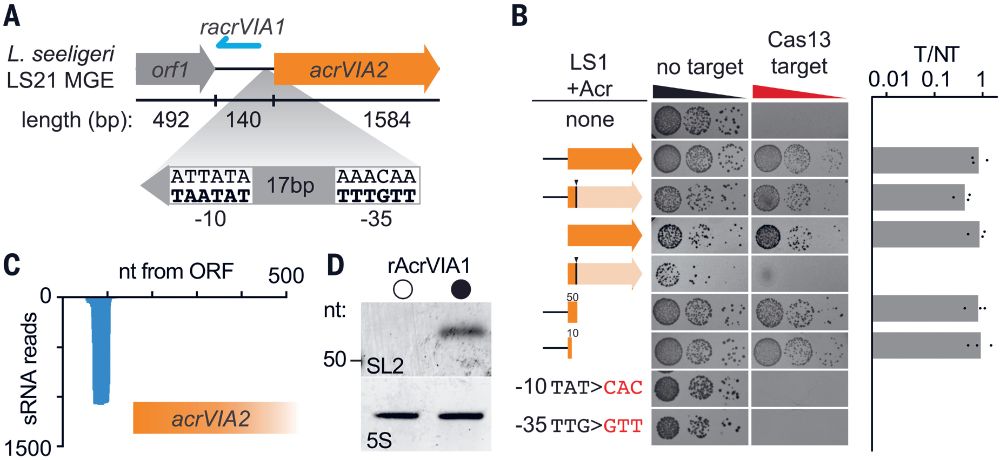
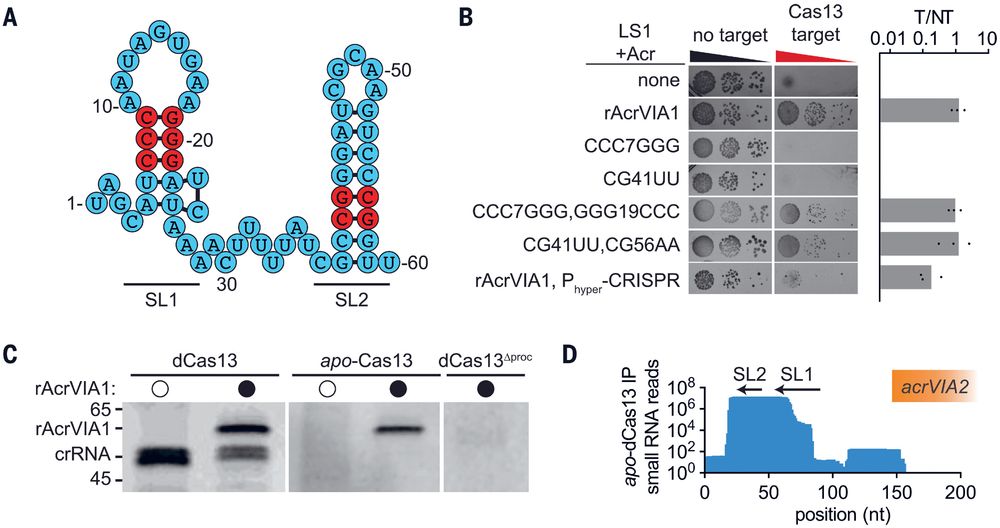
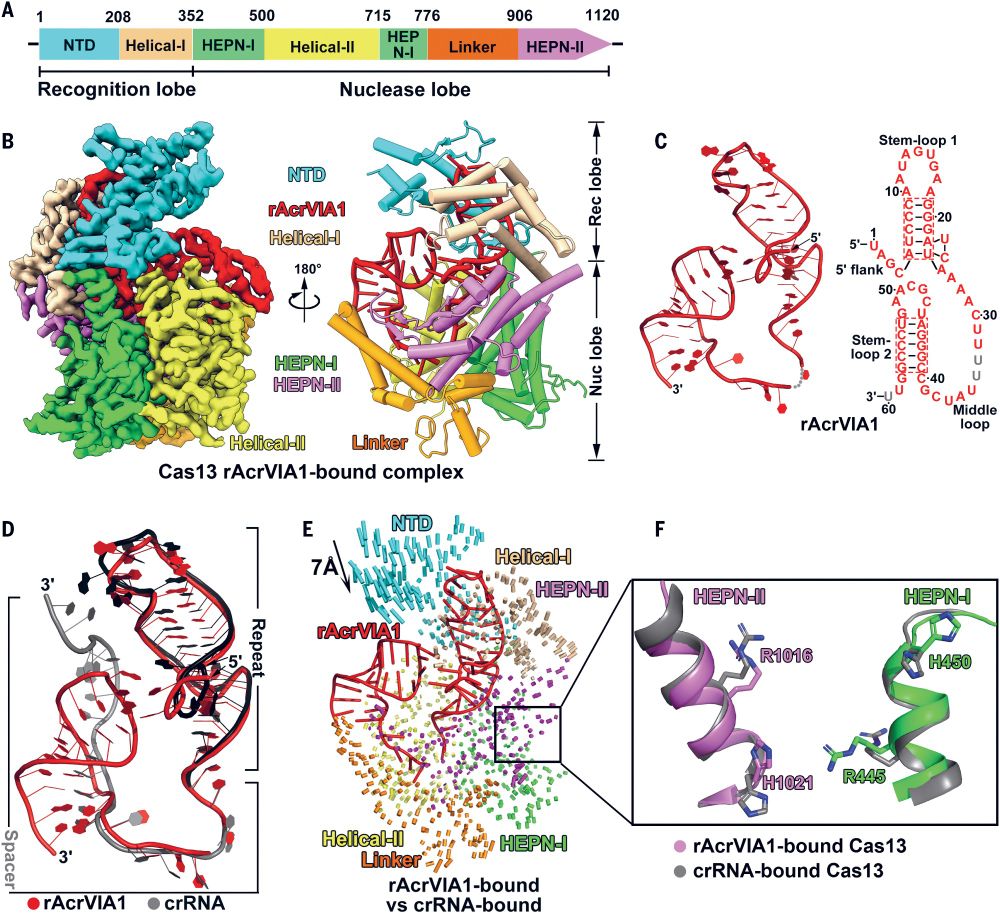
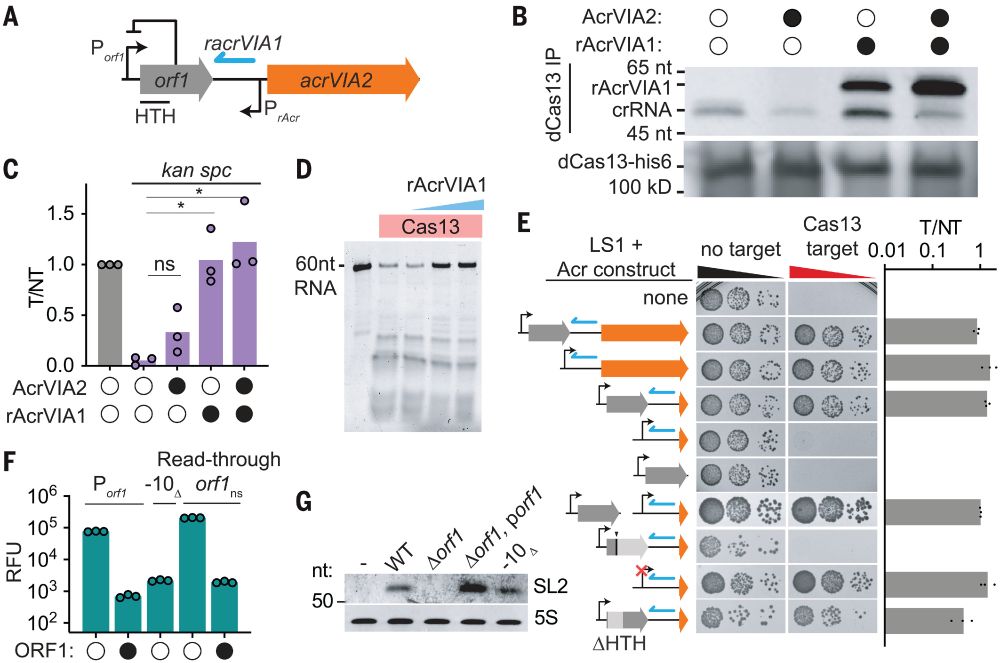
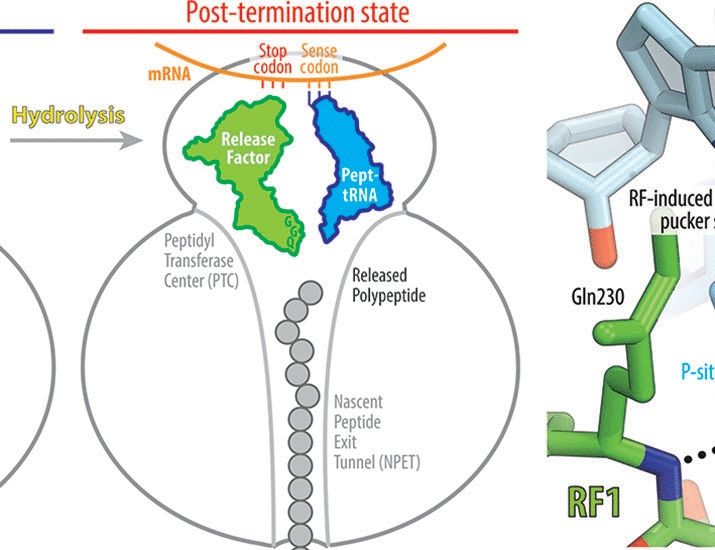
👉https://rdcu.be/eJqFc
bit.ly/46BPXtJ

👉https://rdcu.be/eJqFc
bit.ly/46BPXtJ



🎙️Oliver Mühlemann opens with 'Deciphering the role of the deubiquitinase USP9X in ribosome-associated quality control' at #EMBLProtein.


🎙️Oliver Mühlemann opens with 'Deciphering the role of the deubiquitinase USP9X in ribosome-associated quality control' at #EMBLProtein.
Kicking off our first conference after the summer break: 'Protein synthesis and translational control'.
🌟Today’s highlights include keynote lectures from Lori Passmore, Reuven Agami, and Marina Rodnina.
Welcome to #EMBLProtein! 🎉


Kicking off our first conference after the summer break: 'Protein synthesis and translational control'.
🌟Today’s highlights include keynote lectures from Lori Passmore, Reuven Agami, and Marina Rodnina.
Welcome to #EMBLProtein! 🎉
I’m thrilled to share our new paper (Wolin et al., Cell 2025).
This paper describes SPIDR, a high-throughput method for mapping RBP binding sites.
By combining #SPIDR with #cryoEM, we identified the exact binding site of LARP1 within the #mRNA channel of the 40S ribosomal subunit.
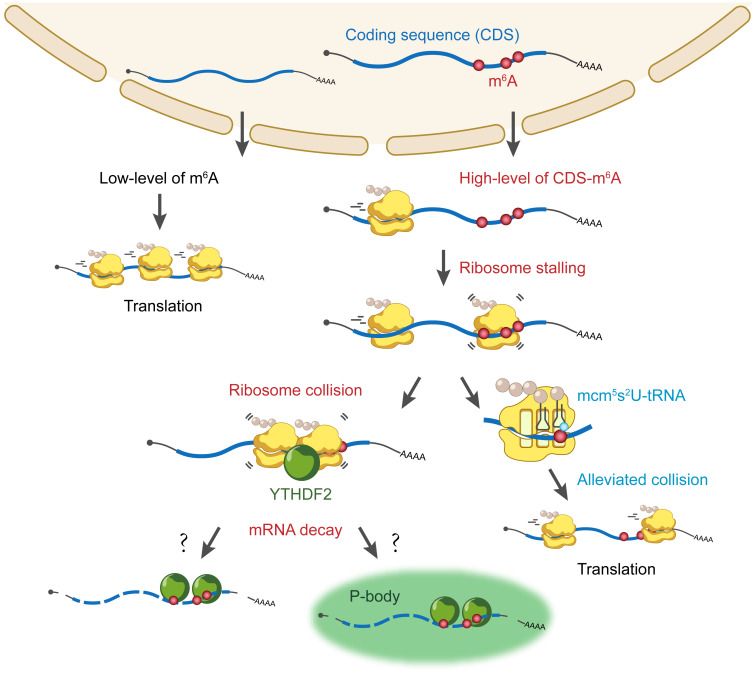
Haleigh Wooters, Neil Nimmagadda, Alicia Darnell, and @thesilvalab.bsky.social .
#Ubiquitination #TranslationalReprogramming #StressResponse
Read it here: authors.elsevier.com/a/1lPPS3S6Gf...

Haleigh Wooters, Neil Nimmagadda, Alicia Darnell, and @thesilvalab.bsky.social .
#Ubiquitination #TranslationalReprogramming #StressResponse
Read it here: authors.elsevier.com/a/1lPPS3S6Gf...
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...
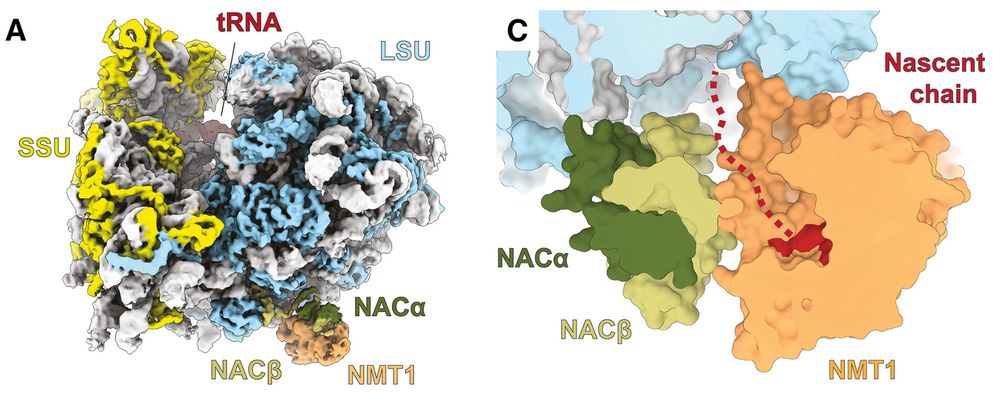
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...
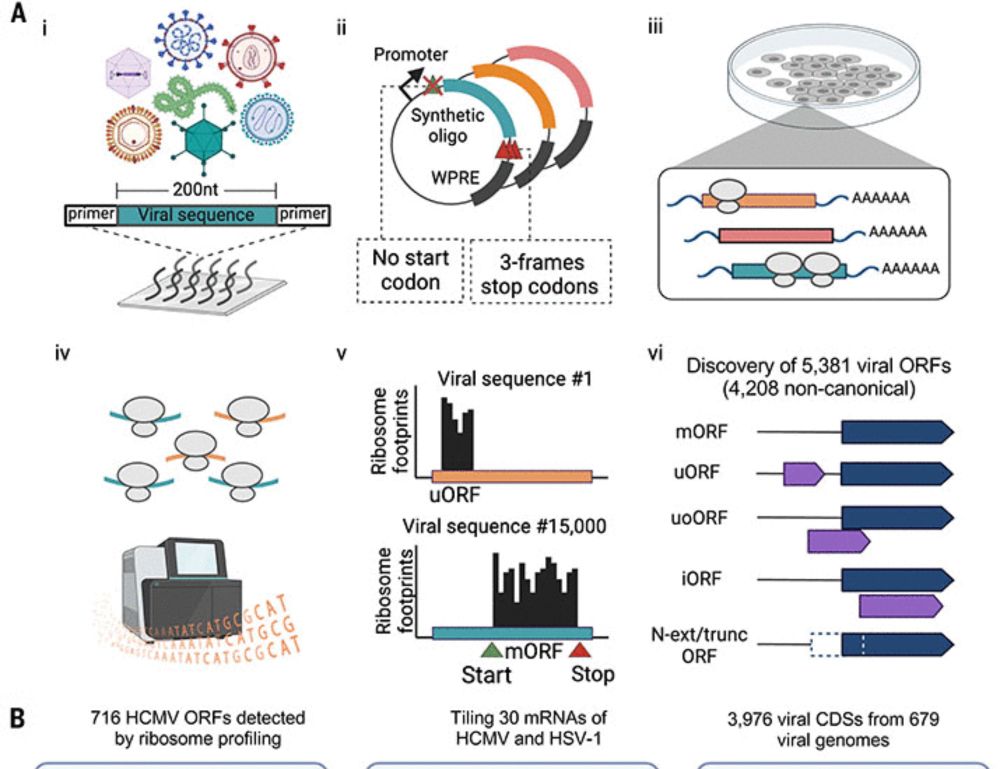
Abstracts to present are due 12 June, and registration closes 22 August.
Find more info here: tinyurl.com/ribosomesynt...

Abstracts to present are due 12 June, and registration closes 22 August.
Find more info here: tinyurl.com/ribosomesynt...

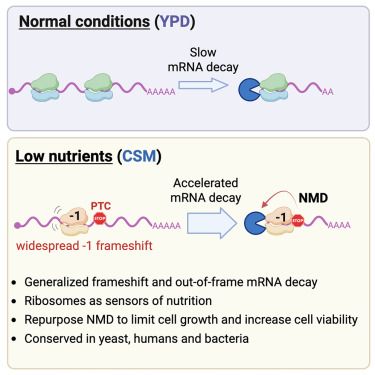

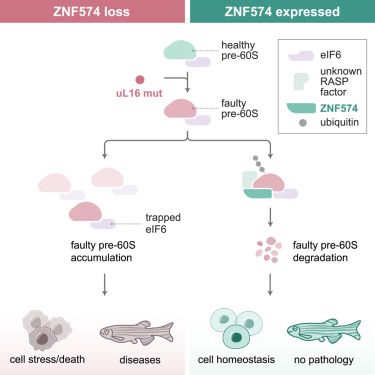
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.cell.com/cell/abstrac...

www.cell.com/cell/abstrac...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


Huge thanks to @ppenev.bsky.social @Amos Nissley @Dipti Nayak @Rohan Sachdeva @jhdcate.bsky.social @Jill Banfield @banfieldlab.bsky.social !!