

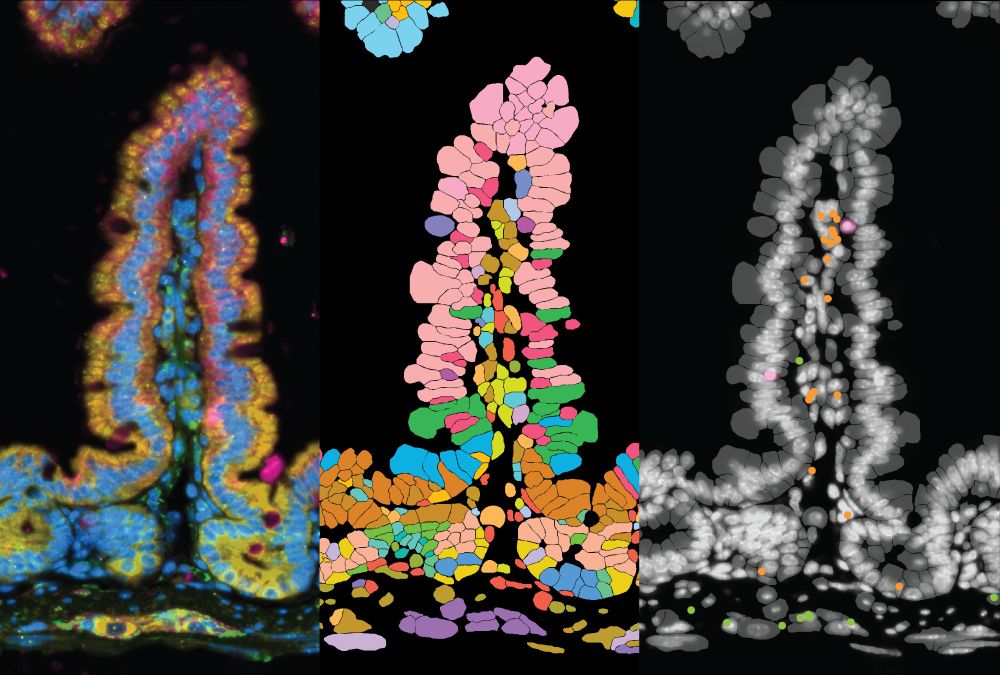

www.nature.com/articles/s41...
Spatial transcriptomics methods have been slow to move into clinical practice, but spatial proteomics are cheaper and more scalable, and could progress faster go.nature.com/3PoWPkz

Spatial transcriptomics methods have been slow to move into clinical practice, but spatial proteomics are cheaper and more scalable, and could progress faster go.nature.com/3PoWPkz
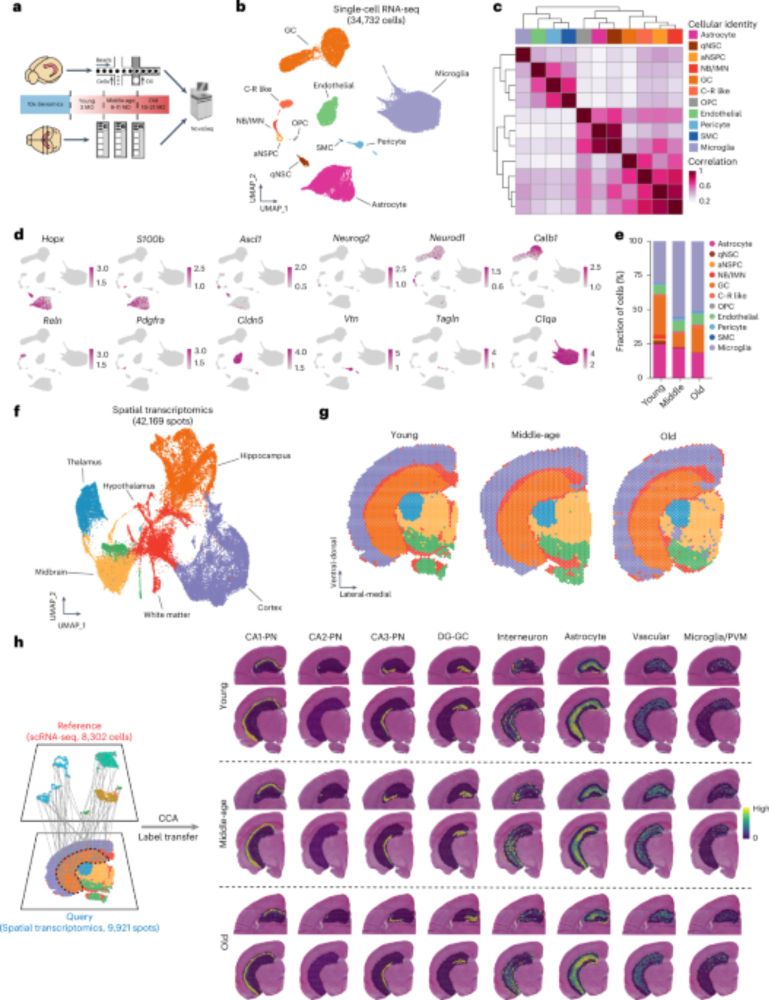
www.nature.com/articles/s42...

www.nature.com/articles/s42...
Erika Paolini used spatial transcriptomics to map the metabolic zonation of hepatocytes in MASLD, showing that PNPLA3 affects zonation by impairing periportal function
🧫🔬⚗️🧬🧪
@easlnews.bsky.social #liversky
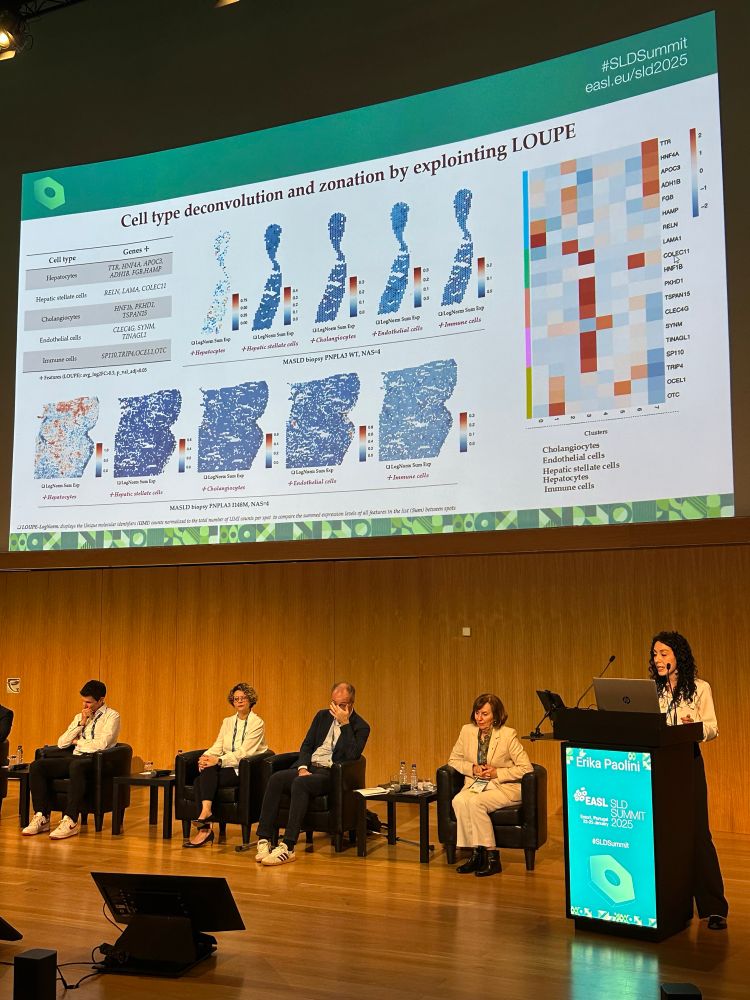
Erika Paolini used spatial transcriptomics to map the metabolic zonation of hepatocytes in MASLD, showing that PNPLA3 affects zonation by impairing periportal function
🧫🔬⚗️🧬🧪
@easlnews.bsky.social #liversky

Highly multiplexed spatial transcriptomics in bacteria | Science www.science.org/doi/10.1126/...
#microsky #microbial #microbiology #genomics
#transcriptomic 🦠🧫

Highly multiplexed spatial transcriptomics in bacteria | Science www.science.org/doi/10.1126/...
#microsky #microbial #microbiology #genomics
#transcriptomic 🦠🧫

