
Researcher @ University of Oslo

We sampled #klebsiella isolates from💩 patients when they entered the hospital & 🩸 after development of infection.
🎯 We tracked phenotypic changes & correlate them w genotypic changes.
@klebclub.bsky.social
-in collab with Clermont-Ferrand lab
#microsky
🧵
doi.org/10.64898/202...

We sampled #klebsiella isolates from💩 patients when they entered the hospital & 🩸 after development of infection.
🎯 We tracked phenotypic changes & correlate them w genotypic changes.
@klebclub.bsky.social
-in collab with Clermont-Ferrand lab
#microsky
🧵
doi.org/10.64898/202...

@martinsteinegger.bsky.social andco
@martinsteinegger.bsky.social andco
bktitanji.substack.com/p/how-unethi...

bktitanji.substack.com/p/how-unethi...
With @tmthrz.bsky.social and @rayanchikhi.bsky.social we aim to tackle practical unitigs compression!
A thread:
With @tmthrz.bsky.social and @rayanchikhi.bsky.social we aim to tackle practical unitigs compression!
A thread:
Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...
Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...
www.the-scientist.com/the-ice-is-a...

www.the-scientist.com/the-ice-is-a...

k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
Full article available: https://doi.org/10.1093/bioadv/vbaf290
Authors include: @pavelvesely.bsky.social, @brinda.eu

k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?
👉 www.biorxiv.org/content/10.6...

👉 www.biorxiv.org/content/10.6...
We show how Lamassu antiphage system, originated from a DNA-repair complex and evolved into a compact and modular immune machine, wt Dinshaw Patel lab in @pnas.org.
👏 @matthieu-haudiquet.bsky.social, Arpita Chakravarti & all authors!
doi.org/10.1073/pnas...


We show how Lamassu antiphage system, originated from a DNA-repair complex and evolved into a compact and modular immune machine, wt Dinshaw Patel lab in @pnas.org.
👏 @matthieu-haudiquet.bsky.social, Arpita Chakravarti & all authors!
doi.org/10.1073/pnas...
#MicroSky #IDSky #Protistsonsky 🧬💻
www.microbiologyresearch.org/content/micr...
#MicroSky #IDSky #Protistsonsky 🧬💻
www.microbiologyresearch.org/content/micr...
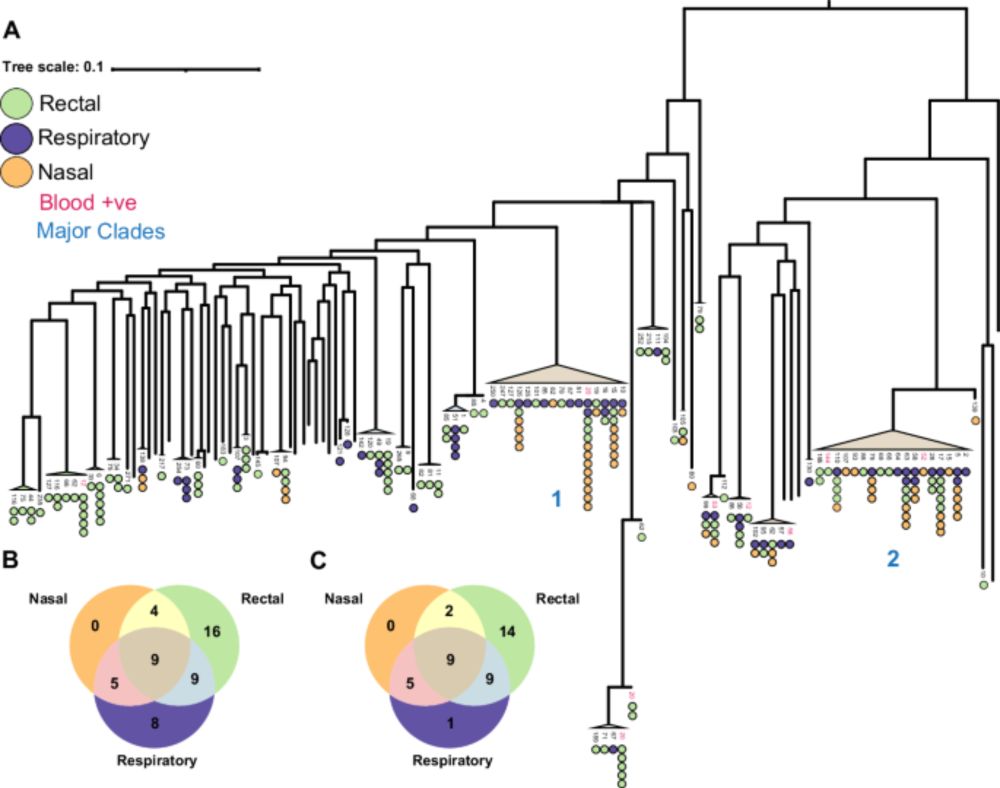
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...
Anyone who’s tried deleting prophages in the lab by HR knows the difficulties of the task. Here we have an example of how HR-mediated natural transformation might hit the same hurdle in a more native context.
academic.oup.com/mbe/article/...

Anyone who’s tried deleting prophages in the lab by HR knows the difficulties of the task. Here we have an example of how HR-mediated natural transformation might hit the same hurdle in a more native context.
academic.oup.com/mbe/article/...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
journals.plos.org/plosbiology/...

journals.plos.org/plosbiology/...
We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use
EMBL-EBI’s new AMR portal brings together laboratory resistance data and bacterial genomes in one open platform.
#WAAW2025 #ActOnAMR
www.ebi.ac.uk/about/news/t...
🧬💻

We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

