
🔹 Want to join? Register here > forms.gle/XWTd4gJAXZSx...
🚨 Limited spots available! So first come, first served!
🔹 More info > crolllab.github.io/ripm2025/
This work has been a major project of @tbadet.bsky.social during his time in our lab.
We hope to stimulate much more exciting work on fungal TEs.
journals.plos.org/plosbiology/...
This work has been a major project of @tbadet.bsky.social during his time in our lab.
We hope to stimulate much more exciting work on fungal TEs.
journals.plos.org/plosbiology/...

@i2bcparissaclay.bsky.social @univparissaclay.bsky.social
@i2bcparissaclay.bsky.social @univparissaclay.bsky.social
Transposable elements (TEs) don't just jump within fungal genomes, they also move extensively between species. In this study, we screened over 1,300 fungal genomes and found a conservative estimate of 5,500+ horizontal transfer events.
Transposable elements (TEs) don't just jump within fungal genomes, they also move extensively between species. In this study, we screened over 1,300 fungal genomes and found a conservative estimate of 5,500+ horizontal transfer events.
We explored the 3D genome of Zymo and found:
🧬 Rabl-like conformation
🧩 Accessory chr tethered to core centromeres
🏰 Self-interacting domains and boundaries with insulator motifs
www.biorxiv.org/content/10.1...
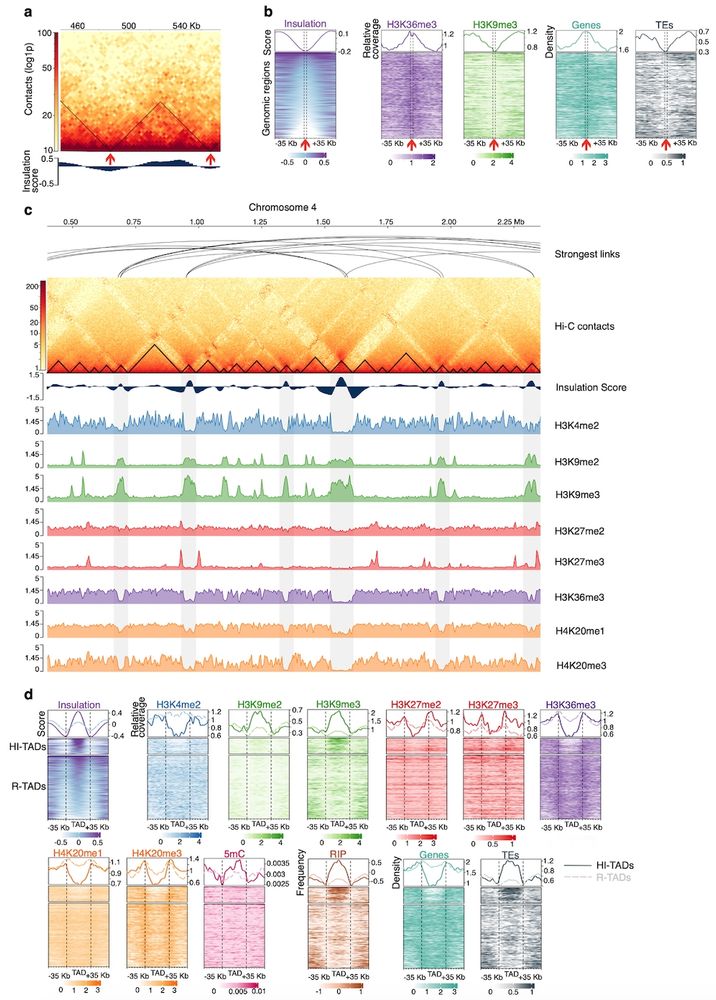
We explored the 3D genome of Zymo and found:
🧬 Rabl-like conformation
🧩 Accessory chr tethered to core centromeres
🏰 Self-interacting domains and boundaries with insulator motifs
www.biorxiv.org/content/10.1...

ℹ️ Deadline is set to May 23rd.
🔥 Many slots are still available.
🔹 Want to join? Register here > forms.gle/XWTd4gJAXZSx...
🚨 Limited spots available! So first come, first served!
🔹 More info > crolllab.github.io/ripm2025/
ℹ️ Deadline is set to May 23rd.
🔥 Many slots are still available.
🔹 Want to join? Register here > forms.gle/XWTd4gJAXZSx...
🚨 Limited spots available! So first come, first served!
🔹 More info > crolllab.github.io/ripm2025/
🔹 Want to join? Register here > forms.gle/XWTd4gJAXZSx...
🚨 Limited spots available! So first come, first served!
🔹 More info > crolllab.github.io/ripm2025/
Resistance in agriculture emerges fast, but we fail to grasp how the myriad mutations in pathogen populations contribute to this.
Please check out and share our latest preprint: www.biorxiv.org/content/10.1...
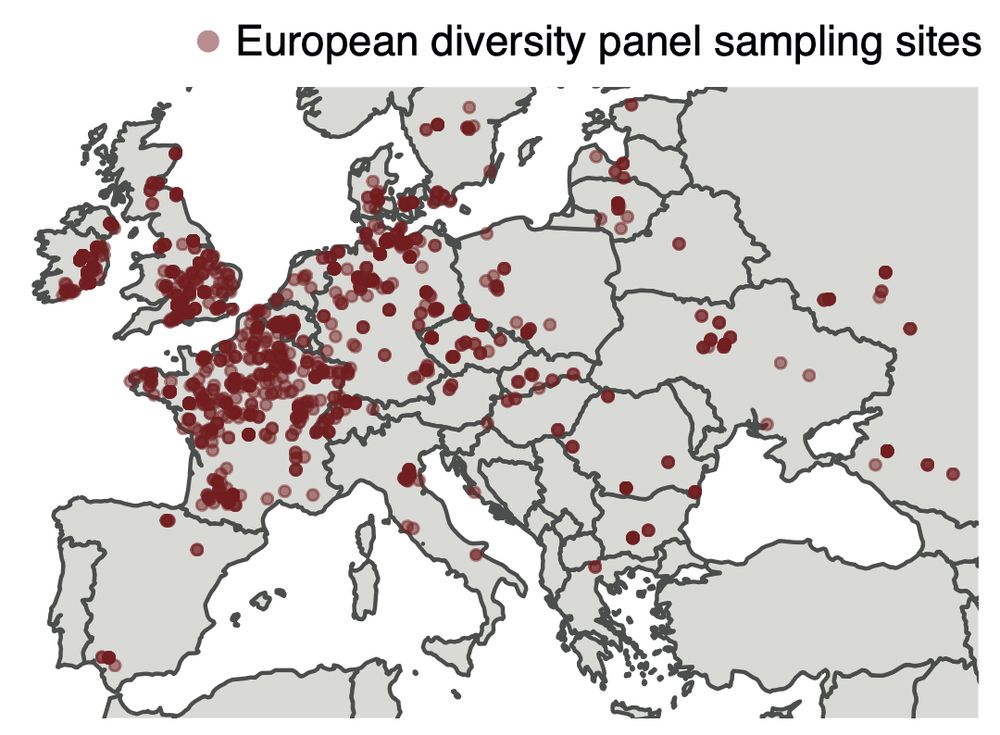
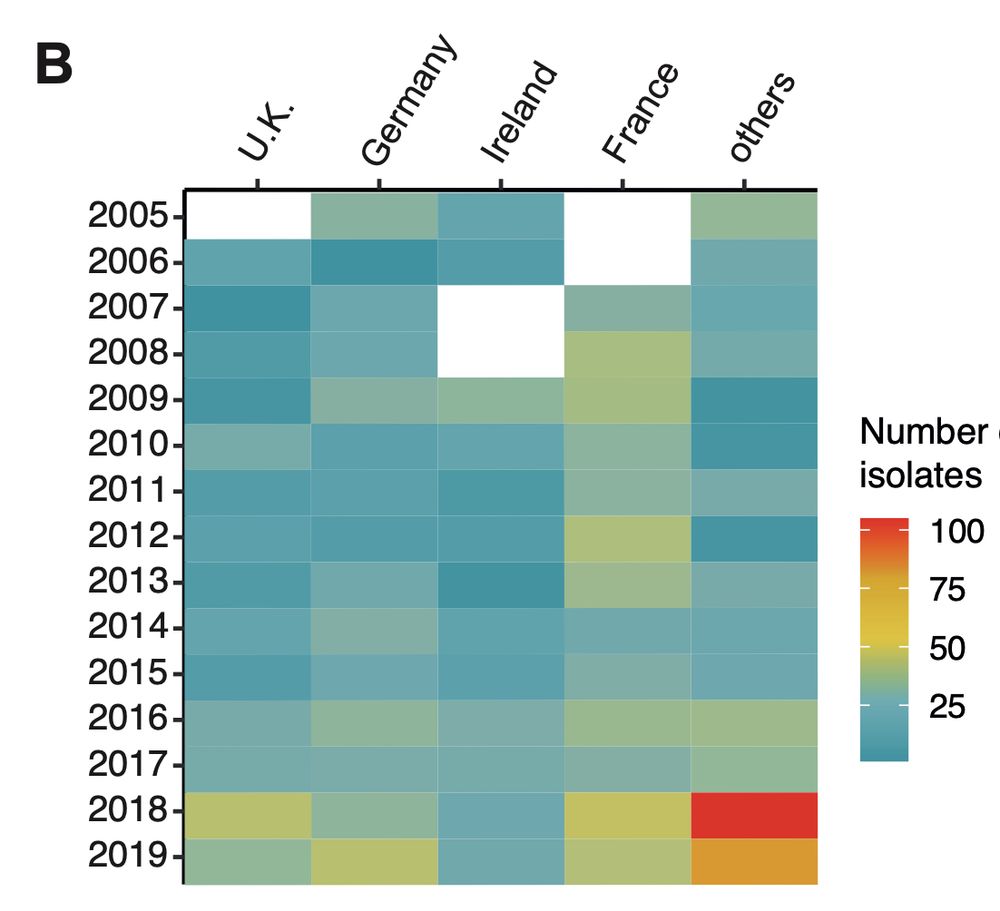
Resistance in agriculture emerges fast, but we fail to grasp how the myriad mutations in pathogen populations contribute to this.
Please check out and share our latest preprint: www.biorxiv.org/content/10.1...
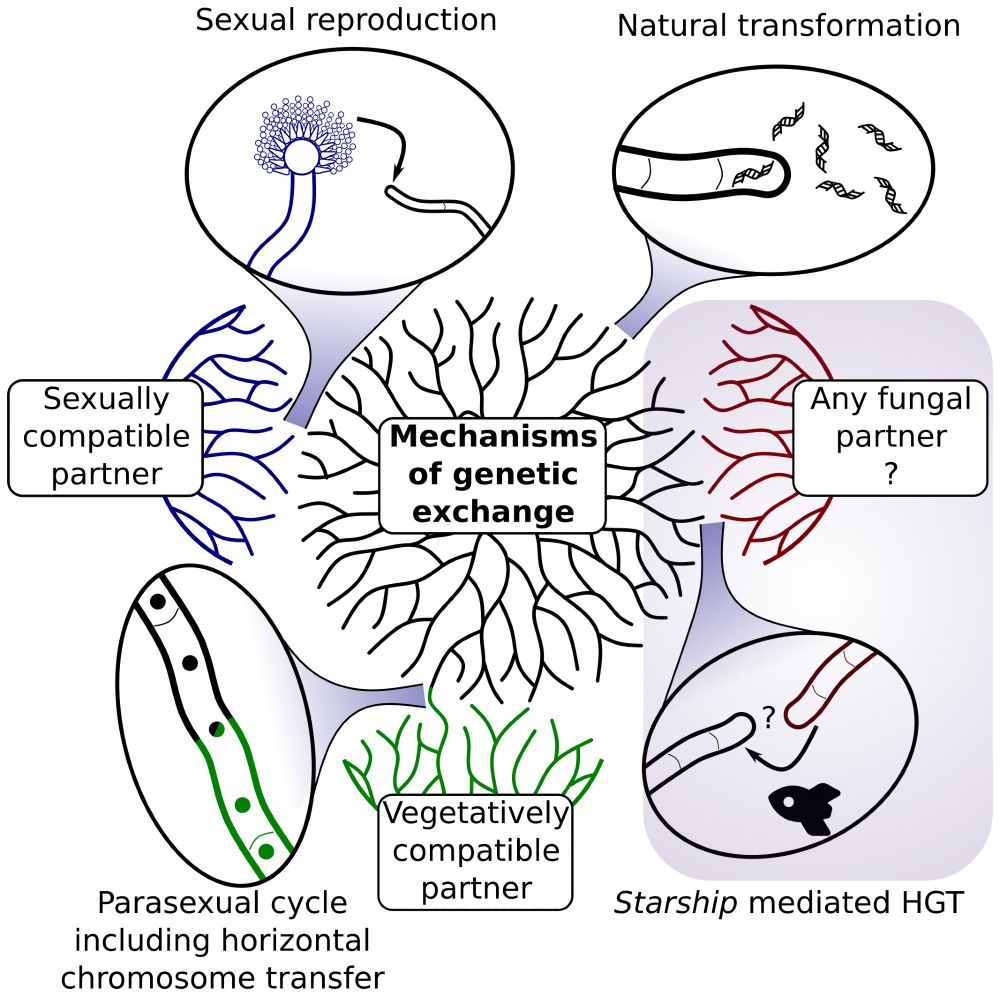
#ecfg17

#ecfg17
🖥️🧬 🧪
In the present paper, our PhD student Nina Marthe developed a Python tool, a user friendly one, that allows to
1/5
www.biorxiv.org/content/10.1...
🖥️🧬 🧪
In the present paper, our PhD student Nina Marthe developed a Python tool, a user friendly one, that allows to
1/5
www.biorxiv.org/content/10.1...

doi.org/10.1093/molb...
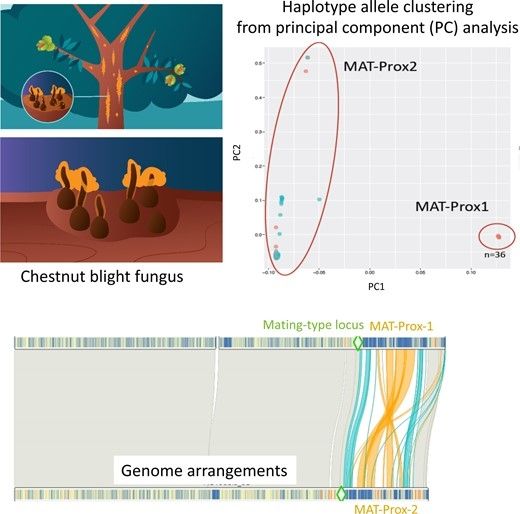
doi.org/10.1093/molb...
Genome size evolution is fascinating and showcases the dynamic nature of transposon activation.
What we understand less are the dynamics of transposon defense systems encoded by the genomes.
doi.org/10.1101/2025...
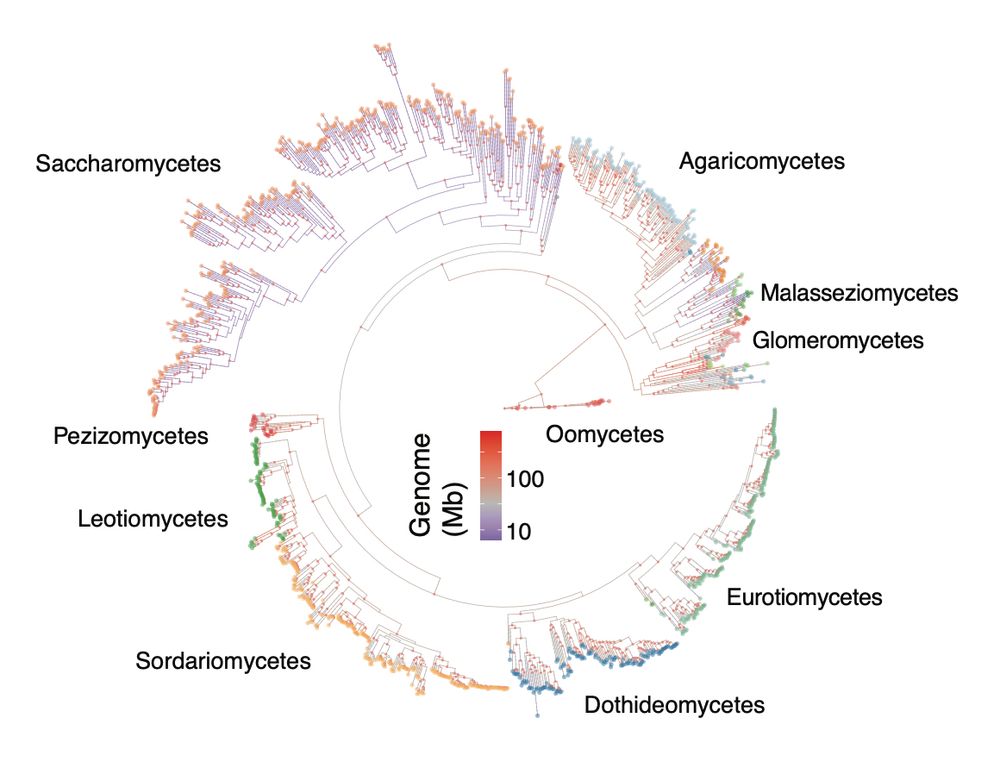
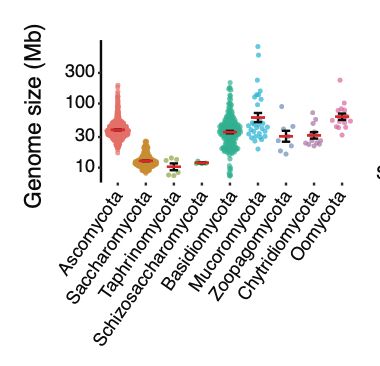
Very interesting results.
Genome size evolution is fascinating and showcases the dynamic nature of transposon activation.
What we understand less are the dynamics of transposon defense systems encoded by the genomes.
doi.org/10.1101/2025...


Very interesting results.
Genome size evolution is fascinating and showcases the dynamic nature of transposon activation.
What we understand less are the dynamics of transposon defense systems encoded by the genomes.
doi.org/10.1101/2025...


Genome size evolution is fascinating and showcases the dynamic nature of transposon activation.
What we understand less are the dynamics of transposon defense systems encoded by the genomes.
doi.org/10.1101/2025...

