
lorrainlab.owlstown.net
In this review, Lee & Seo highlight the hierarchical organization of plant genomes, emphasizing recent advances in understanding local chromatin structures shaped by cohesins and gene borders as fundamental structural units.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

In this review, Lee & Seo highlight the hierarchical organization of plant genomes, emphasizing recent advances in understanding local chromatin structures shaped by cohesins and gene borders as fundamental structural units.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
www.nature.com/articles/s41...

www.nature.com/articles/s41...
🌱Check out our new Opinion Paper with @thanvisrikant.bsky.social discussing this topic in @cp-trendsplantsci.bsky.social!
www.sciencedirect.com/science/arti...

🌱Check out our new Opinion Paper with @thanvisrikant.bsky.social discussing this topic in @cp-trendsplantsci.bsky.social!
www.sciencedirect.com/science/arti...
Paradigm shift ahead! We discovered a new mode of DNA methylation targeting in plants that relies on transcription factors and sequence motifs rather than chromatin modifications to regulate the methylome. rdcu.be/eQ6L5
1/8

Paradigm shift ahead! We discovered a new mode of DNA methylation targeting in plants that relies on transcription factors and sequence motifs rather than chromatin modifications to regulate the methylome. rdcu.be/eQ6L5
1/8

The Expanding Histone Universe: Histone-Based DNA Organization in Noneukaryotic Organisms - www.annualreviews.org/content/jour...

The Expanding Histone Universe: Histone-Based DNA Organization in Noneukaryotic Organisms - www.annualreviews.org/content/jour...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
@currentbiology.bsky.social by @jromeijn.bsky.social et al from @mfseidl.bsky.social
www.cell.com/current-biol...

@currentbiology.bsky.social by @jromeijn.bsky.social et al from @mfseidl.bsky.social
www.cell.com/current-biol...
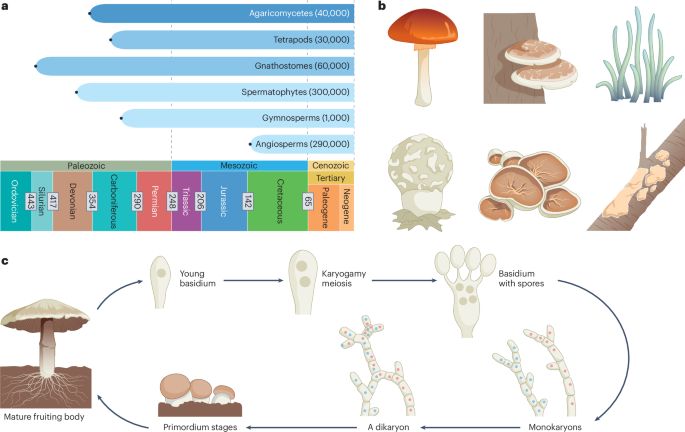
Find out in our new paper by Alger Jorritsma, where we show which strategies evolve under different environmental conditions. @journal-evo.bsky.social
academic.oup.com/evolut/advan...

Find out in our new paper by Alger Jorritsma, where we show which strategies evolve under different environmental conditions. @journal-evo.bsky.social
academic.oup.com/evolut/advan...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
link.springer.com/article/10.1...

link.springer.com/article/10.1...
@natsmb.nature.com. A wonderful team effort across the centromere community, across @jansenlab.bsky.social @naltemose.bsky.social @dfachinetti.bsky.social and Giunta labs. Happy reading! 1/4
www.nature.com/articles/s41...

@natsmb.nature.com. A wonderful team effort across the centromere community, across @jansenlab.bsky.social @naltemose.bsky.social @dfachinetti.bsky.social and Giunta labs. Happy reading! 1/4
www.nature.com/articles/s41...

No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

go.nature.com/47Fsqax

go.nature.com/47Fsqax
GWAS identifies CDCA7 as a regulator of DNA methylation in Arabidopsis thaliana . CDCA7 binds the chromatin remodeller DDM1 and modulates the control of CG methylation.

GWAS identifies CDCA7 as a regulator of DNA methylation in Arabidopsis thaliana . CDCA7 binds the chromatin remodeller DDM1 and modulates the control of CG methylation.



