
Studying post transcriptional regulation in single cells and Alzheimer's Disease. iPSCs, gene regulatory networks, bioinformatics, systems biology
https://www.plasslab.com/
Our lab opens 2 PhD positions to investigate fungi's evolution: #Barcelona
Wet lab in Biochemistry/Structural Biology of cryo-tolerant fungi
www.upf.edu/documents/d/...
Computational in Physical Chemistry of cellular processes under extrem environments
www.upf.edu/documents/d/...
Our lab opens 2 PhD positions to investigate fungi's evolution: #Barcelona
Wet lab in Biochemistry/Structural Biology of cryo-tolerant fungi
www.upf.edu/documents/d/...
Computational in Physical Chemistry of cellular processes under extrem environments
www.upf.edu/documents/d/...

And the paper certainly reflects all of that. Go check it out!
www.nature.com/articles/s41...

And the paper certainly reflects all of that. Go check it out!
www.nature.com/articles/s41...

www.nature.com/articles/s41...





Join my lab @ub.edu in Barcelona!
We are looking for a junior postdoc to apply for a Juan de la Cierva fellowship. Both computational and hybrid candidates are welcomed! Get in touch to discuss the project & details
Join my lab @ub.edu in Barcelona!
We are looking for a junior postdoc to apply for a Juan de la Cierva fellowship. Both computational and hybrid candidates are welcomed! Get in touch to discuss the project & details

Dona ahora en anticiparsealalzheimer.org

Dona ahora en anticiparsealalzheimer.org
Join my lab @ub.edu in Barcelona!
We are looking for a junior postdoc to apply for a Juan de la Cierva fellowship. Both computational and hybrid candidates are welcomed! Get in touch to discuss the project & details
Join my lab @ub.edu in Barcelona!
We are looking for a junior postdoc to apply for a Juan de la Cierva fellowship. Both computational and hybrid candidates are welcomed! Get in touch to discuss the project & details
📅 Starting date early 2026, with some flexibility.
🙏Please, RT!

📅 Starting date early 2026, with some flexibility.
🙏Please, RT!
Congratulations Dr. Ake @franzake.bsky.social !!! 👏👏👏 🎉🎉🎉
It has been a pleasure to have you in the lab. #PhD_defense #new_PhD

Congratulations Dr. Ake @franzake.bsky.social !!! 👏👏👏 🎉🎉🎉
It has been a pleasure to have you in the lab. #PhD_defense #new_PhD
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
rdcu.be/ezGqT
Work led by @sobonnal.bsky.social, with Simon Bajew & @rmartinezcorral.bsky.social. @crg.eu @upf.edu

rdcu.be/ezGqT
Work led by @sobonnal.bsky.social, with Simon Bajew & @rmartinezcorral.bsky.social. @crg.eu @upf.edu
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
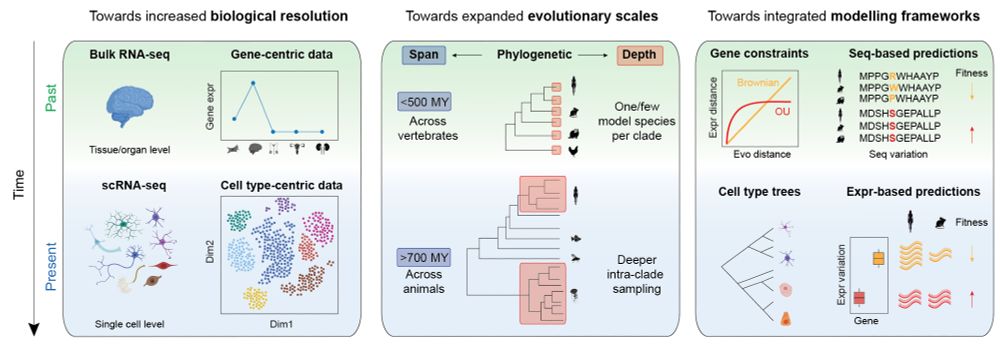
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
👉 humantechnopole.it/en/news/call... #PhDlife

👉 humantechnopole.it/en/news/call... #PhDlife


