🔗 Project details:
www.protaiomics.eu/project/dc12...
👩🔬 Meet the PI:
www.protaiomics.eu/supervisor/v...
👉 Apply here:
www.protaiomics.eu/call-for-app...
#PhDposition #MSCA #Bioinformatics #Proteomics #ArtificialIntelligence #DoctoralTraining #ResearchCareers

🔗 Project details:
www.protaiomics.eu/project/dc12...
👩🔬 Meet the PI:
www.protaiomics.eu/supervisor/v...
👉 Apply here:
www.protaiomics.eu/call-for-app...
#PhDposition #MSCA #Bioinformatics #Proteomics #ArtificialIntelligence #DoctoralTraining #ResearchCareers
@lukaskall.bsky.social, Director of the program, explains how the initiative is designed to identify talent and support researchers who want to build long-term careers in Sweden. ↓
www.scilifelab.se/news/intervi...

@lukaskall.bsky.social, Director of the program, explains how the initiative is designed to identify talent and support researchers who want to build long-term careers in Sweden. ↓
www.scilifelab.se/news/intervi...
If you want to work at the intersection of AI and proteomics, with international training and mobility across Europe, this is your moment.Deadline: 30 November
👉Apply now: www.protaiomics.eu
#Proteomics #AI #MSCA #PhDOpportunities

If you want to work at the intersection of AI and proteomics, with international training and mobility across Europe, this is your moment.Deadline: 30 November
👉Apply now: www.protaiomics.eu
#Proteomics #AI #MSCA #PhDOpportunities
us-hupo.org/Distinguishe...

us-hupo.org/Distinguishe...
us-hupo.org/Computationa...

us-hupo.org/Computationa...
Vaccinations to prevent cervical cancer have plummeted in Britain
www.economist.com/britain/2025...

Vaccinations to prevent cervical cancer have plummeted in Britain
www.economist.com/britain/2025...

pubs.acs.org/doi/10.1021/...
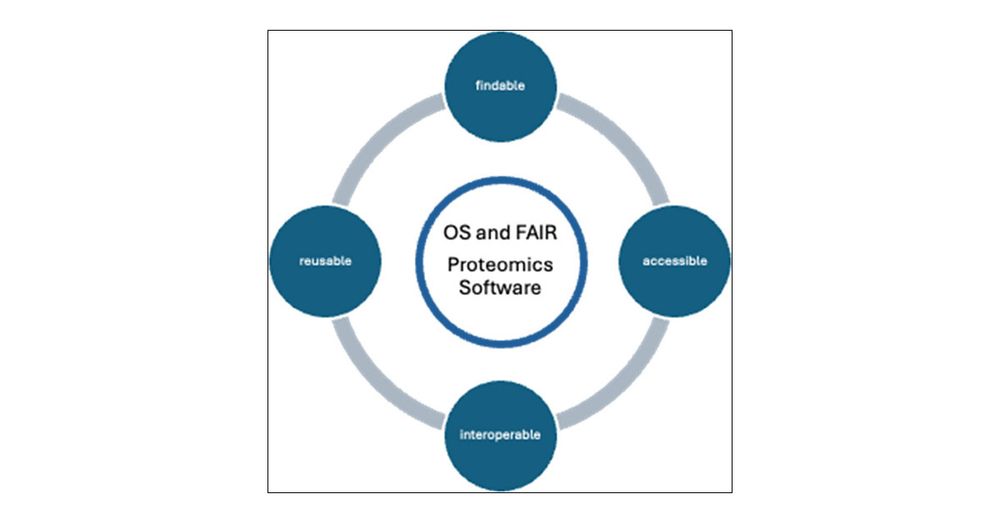
pubs.acs.org/doi/10.1021/...
Join us in Harrachov, Czechia for a week of keynotes, workshops, and networking on computational MS.
Info and registration: eubic-ms.org/events/2026-...
#EuBIC2026 #MassSpectrometry #MassSpec #Bioinformatics #Proteomics #Metabolomics

Join us in Harrachov, Czechia for a week of keynotes, workshops, and networking on computational MS.
Info and registration: eubic-ms.org/events/2026-...
#EuBIC2026 #MassSpectrometry #MassSpec #Bioinformatics #Proteomics #Metabolomics
A human pan-disease🩸 atlas of the circulating proteome
👉 www.proteinatlas.org/humanproteom...
📖 www.science.org/doi/10.1126/...
🤩 @buenoalvez.bsky.social and many more!
@kawresearch.bsky.social
@scilifelab.se
@kthuniversity.bsky.social
A human pan-disease🩸 atlas of the circulating proteome
👉 www.proteinatlas.org/humanproteom...
📖 www.science.org/doi/10.1126/...
🤩 @buenoalvez.bsky.social and many more!
@kawresearch.bsky.social
@scilifelab.se
@kthuniversity.bsky.social
Please check out all the project descriptions at the site below:
Please check out all the project descriptions at the site below:

The ProtAIomics Doctoral Network (ProtAIomics-DN) is launching soon!
💡 We are offering 16 fully funded PhD positions to shape the future of AI-driven proteomics research.
👉 Stay tuned for updates!
#PhD #Proteomics #ArtificialIntelligence #MSCA #HorizonEurope #PhDOpportunities

The ProtAIomics Doctoral Network (ProtAIomics-DN) is launching soon!
💡 We are offering 16 fully funded PhD positions to shape the future of AI-driven proteomics research.
👉 Stay tuned for updates!
#PhD #Proteomics #ArtificialIntelligence #MSCA #HorizonEurope #PhDOpportunities


ESPOD builds on the collaborative relationship between EMBL-EBI and the @sangerinstitute.bsky.social, offering projects that combine experimental and computational approaches.
www.ebi.ac.uk/research/pos...
🧬🖥️🔬#postdocjob

ESPOD builds on the collaborative relationship between EMBL-EBI and the @sangerinstitute.bsky.social, offering projects that combine experimental and computational approaches.
www.ebi.ac.uk/research/pos...
🧬🖥️🔬#postdocjob
www.nordicmsp2025.com

www.nordicmsp2025.com

www.embopress.org/doi/full/10....

www.embopress.org/doi/full/10....



