grants.nih.gov/news-events/...
grants.nih.gov/news-events/...
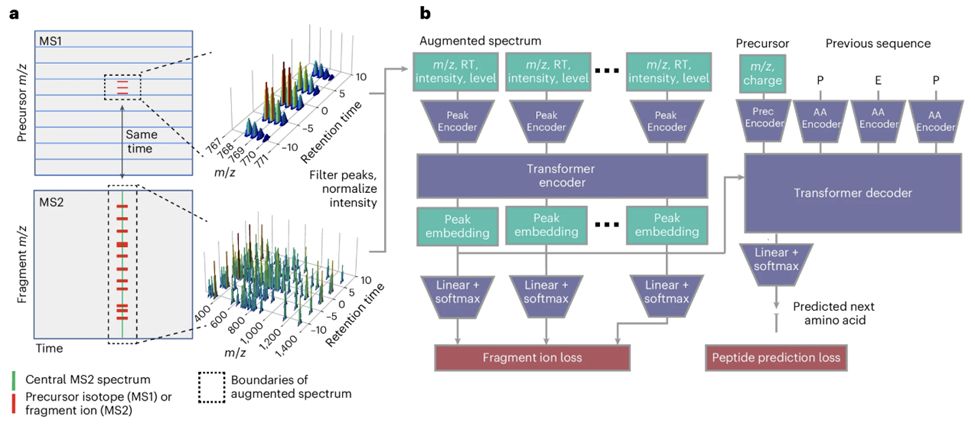
www.nature.com/articles/s41...
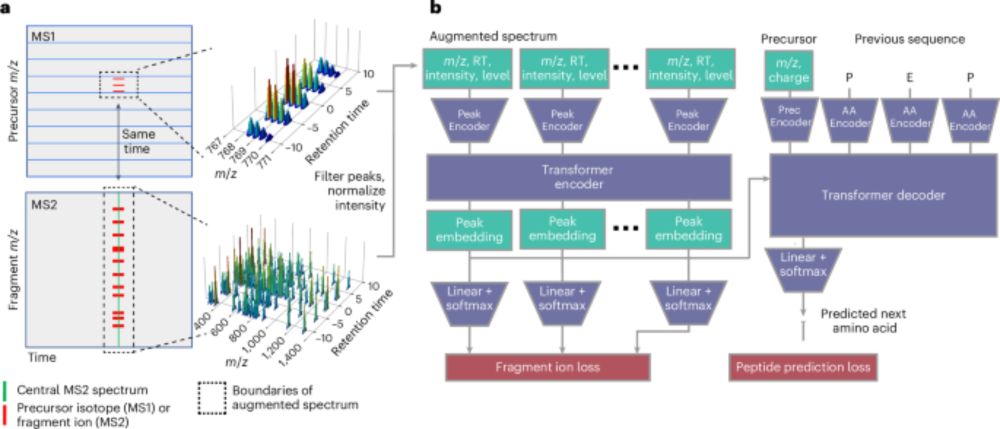
www.nature.com/articles/s41...
www.nature.com/articles/s41...
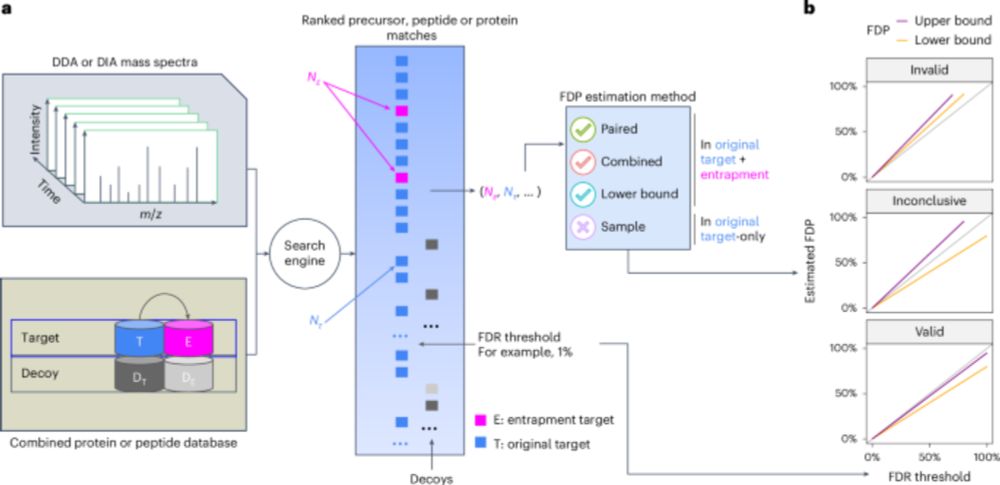
www.nature.com/articles/s41...
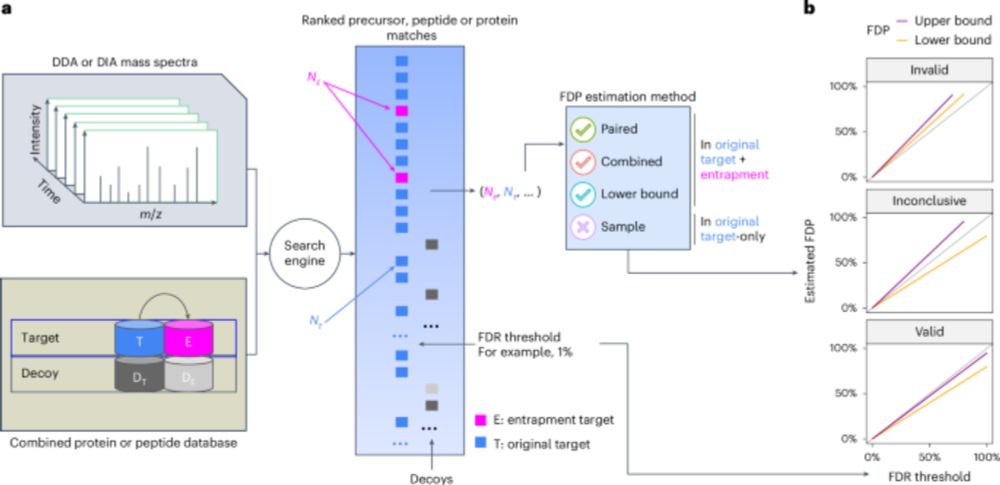
www.nature.com/articles/s41...

Available on GitHub and installable with `pip install ledidi`
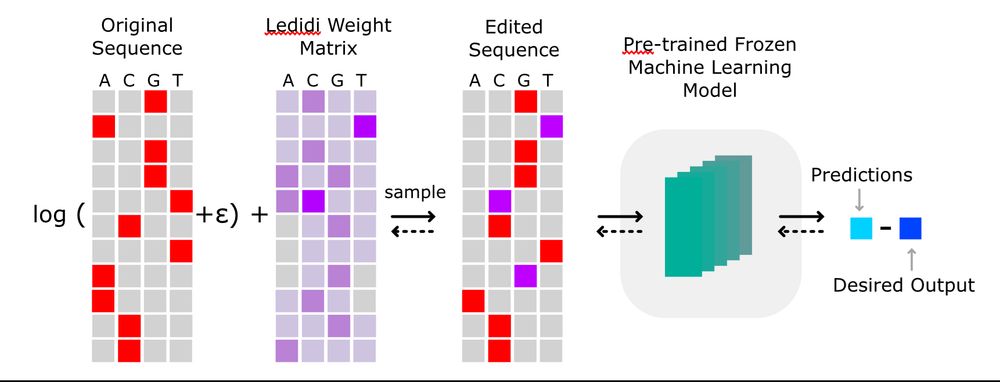
Available on GitHub and installable with `pip install ledidi`

bit.ly/3ZDgYt8

bit.ly/3ZDgYt8
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
How can you transfer peptide IDs between runs and still control your false discovery rate? Till now, the short answer is, you couldn't. Now you can, with PIP-ECHO.
www.biorxiv.org/content/10.1...

How can you transfer peptide IDs between runs and still control your false discovery rate? Till now, the short answer is, you couldn't. Now you can, with PIP-ECHO.
www.biorxiv.org/content/10.1...
communities.springernature.com/posts/wrangl...
communities.springernature.com/posts/wrangl...


