#Review by @carolinamonzo.bsky.social, Tianyuan Liu & @anaconesa.bsky.social @conesalab.bsky.social @csic.es
Free to read here: rdcu.be/efBjP

#Review by @carolinamonzo.bsky.social, Tianyuan Liu & @anaconesa.bsky.social @conesalab.bsky.social @csic.es
Free to read here: rdcu.be/efBjP
www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
www.nature.com/articles/s41...
If you work in human genetics or transcriptomics, do not miss our tweetorial!👇
@guigolab.bsky.social @bsc-cns.bsky.social @crg.eu
www.nature.com/articles/s41...
If you work in human genetics or transcriptomics, do not miss our tweetorial!👇
@guigolab.bsky.social @bsc-cns.bsky.social @crg.eu
Read the news: https://loom.ly/QD3C_Ac




More information available at: 👉 longtrec.eu/VALT/
Meet the #LongReadsTranscriptomics community, connect with experts, and explore the latest advances in this research area.


More information available at: 👉 longtrec.eu/VALT/
Meet the #LongReadsTranscriptomics community, connect with experts, and explore the latest advances in this research area.


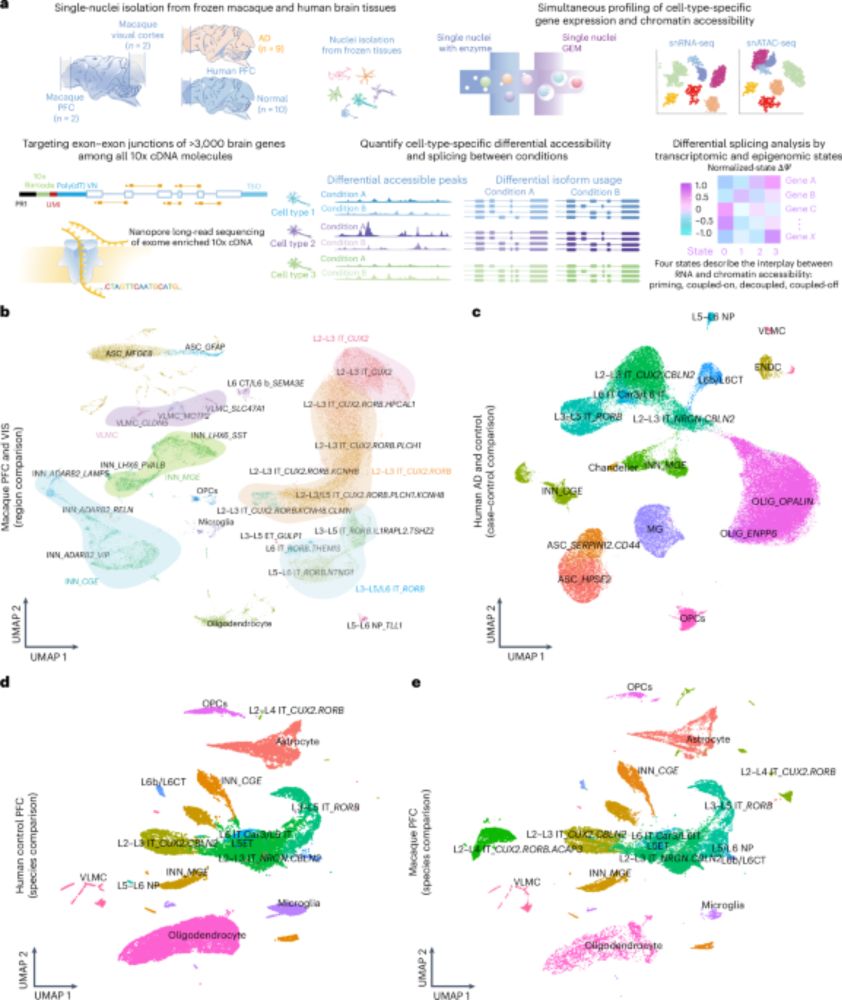
We profiled #RNA isoforms from 31 neuropsychiatric risk genes in the human brain using long-read sequencing. Unannotated isoforms commonly made up a significant proportion of a gene's expression.
genomebiology.biomedcentral.com/articles/10....

We profiled #RNA isoforms from 31 neuropsychiatric risk genes in the human brain using long-read sequencing. Unannotated isoforms commonly made up a significant proportion of a gene's expression.
genomebiology.biomedcentral.com/articles/10....
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
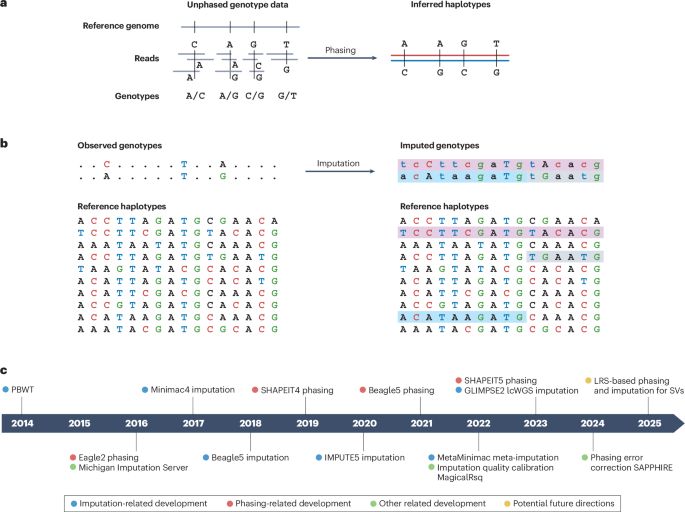
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
conesalab.uv.es/july-from-va...
conesalab.uv.es/july-from-va...




“A systematic benchmark of bioinformatics methods for single-cell and spatial RNA-seq Nanopore long-read data”
🔗 Read here: www.biorxiv.org/content/10.1...
🔬 About the study
We present the first comprehensive benchmark of bioinformatics tools 👇

“A systematic benchmark of bioinformatics methods for single-cell and spatial RNA-seq Nanopore long-read data”
🔗 Read here: www.biorxiv.org/content/10.1...
🔬 About the study
We present the first comprehensive benchmark of bioinformatics tools 👇
@tianyuanliu.bsky.social - "Transcriptome Universal Single-isoform Control (TUSCO) A framework for evaluating transcriptome quality." 15:40–15:50 Track iRNA · Room 02N



@tianyuanliu.bsky.social - "Transcriptome Universal Single-isoform Control (TUSCO) A framework for evaluating transcriptome quality." 15:40–15:50 Track iRNA · Room 02N


We're looking forward to meeting you!!

We're looking forward to meeting you!!