We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K

We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K
in EMBL, which introduced how mammal cells detoxify m6A and how the accumulation influences metabolism. Was wondering how about bacteria. What’s the function of m6A or m6dA in Ecoli. Very in-time preprint 👋
tinyurl.com/ch3damp
We show how molecular byproducts released during virus-induced cell exploitation are used as signals to trigger host immunity
Revealed by the amazing Ilya Osterman. See his thread below👇
in EMBL, which introduced how mammal cells detoxify m6A and how the accumulation influences metabolism. Was wondering how about bacteria. What’s the function of m6A or m6dA in Ecoli. Very in-time preprint 👋
🔗 www.embl.org/news/science...

🔗 www.embl.org/news/science...
@hugovaysset.bsky.social reviewed them all!
Read to know more about all their molecular mechanisms, how phages counteract them, their distribution in bacteria and their conservation in eukaryotic immunity!
www.cell.com/molecular-ce...


www.nature.com/articles/s41...

www.nature.com/articles/s41...

https://go.nature.com/4l27lMN

https://go.nature.com/4l27lMN
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

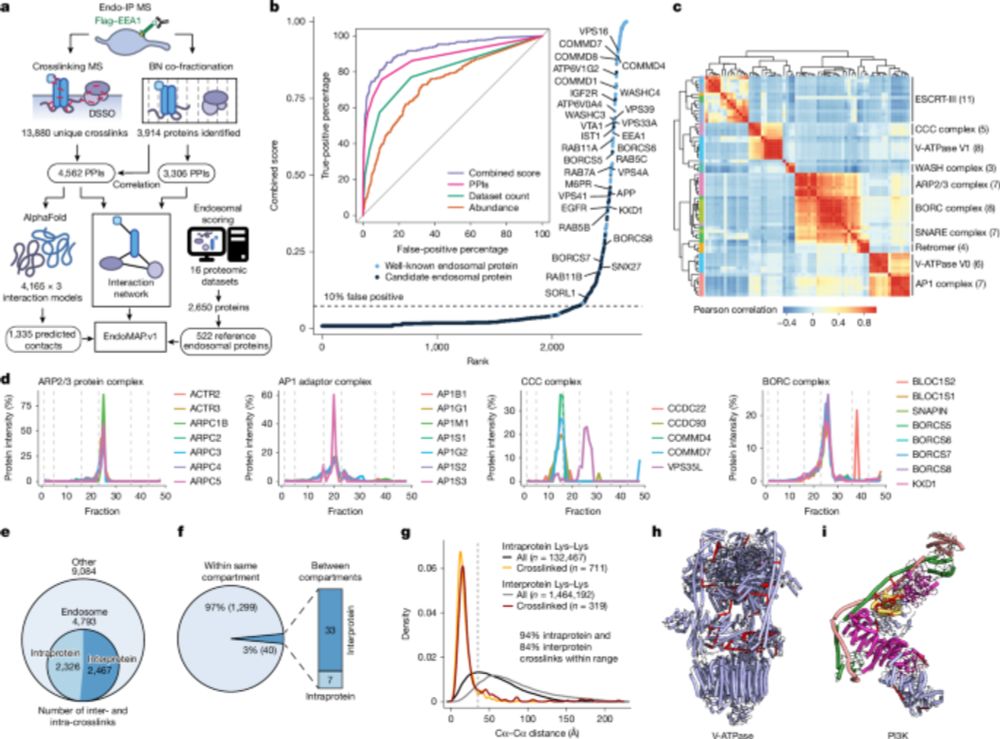
---
#proteomics #prot-preprint

www.nature.com/articles/s41...
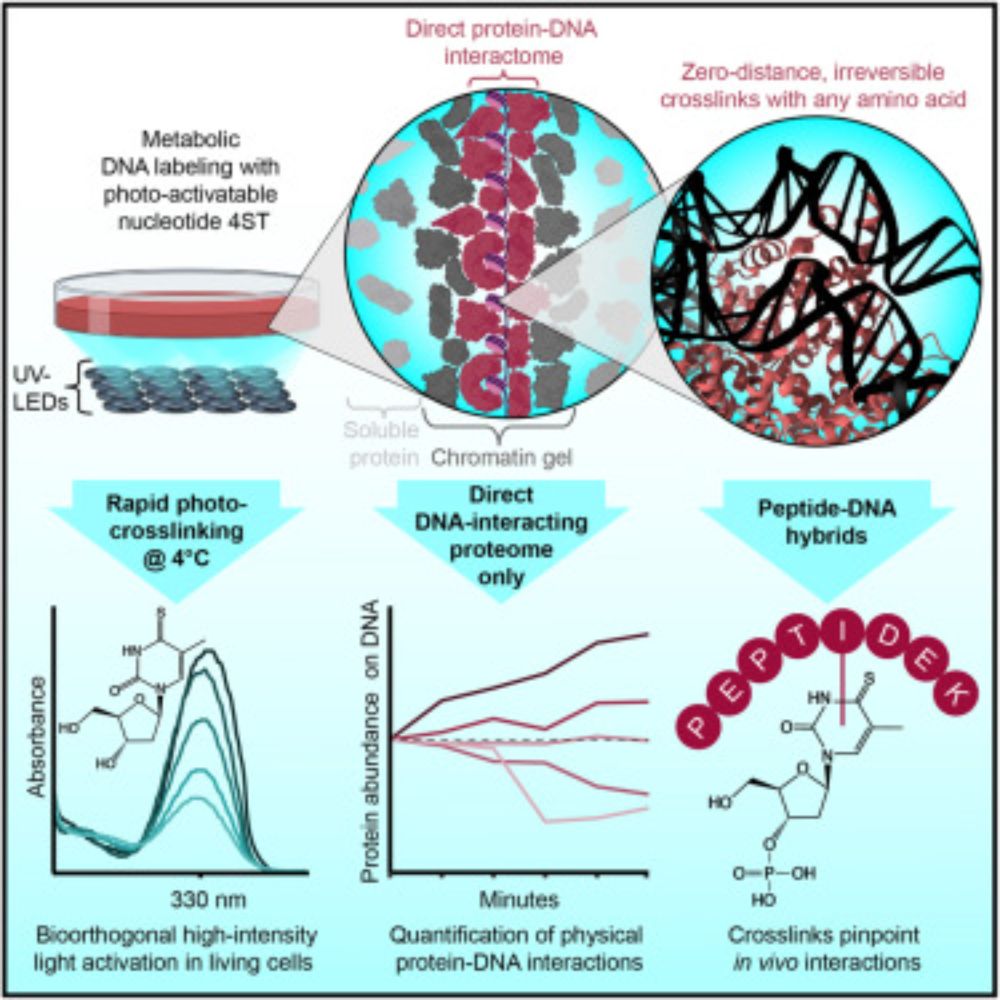
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...


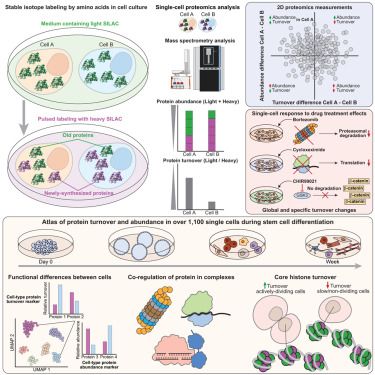
We've developed SC-pSILAC to simultaneously measure protein turnover and abundance in single cells, unlocking the first large-scale, 2D proteomic insights at single-cell resolution!
www.cell.com/cell/fulltex...