
I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:

I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:

I have struggled my way through the academic system, with times when I was about to give up due to lack of funding and support.
I am proud where I am now, and what I have achieved. Thanks to everyone who supported me and believed in me!

I have struggled my way through the academic system, with times when I was about to give up due to lack of funding and support.
I am proud where I am now, and what I have achieved. Thanks to everyone who supported me and believed in me!



www.nature.com/articles/s41...
Congrats to the authors: @sshamphavi.bsky.social @loumollat.bsky.social @mariejoest.bsky.social Najwa Taib and @sgribaldo.bsky.social
www.nature.com/articles/s41...
Congrats to the authors: @sshamphavi.bsky.social @loumollat.bsky.social @mariejoest.bsky.social Najwa Taib and @sgribaldo.bsky.social
@hohlbeinlab.bsky.social @nicoc-micsynmet.bsky.social @ettema.bsky.social @mib-wur.bsky.social
doi.org/10.1038/s414...

@hohlbeinlab.bsky.social @nicoc-micsynmet.bsky.social @ettema.bsky.social @mib-wur.bsky.social
doi.org/10.1038/s414...
Small Things Considered
Big Things Achieved

Small Things Considered
Big Things Achieved
Read the news 👉 https://loom.ly/22XW72k
#ResearchInfrastructure #Science4EU

Read the news 👉 https://loom.ly/22XW72k
#ResearchInfrastructure #Science4EU
GTDB r220 case study (led by @kassipan.bsky.social )
Applied WitChi to the archaeal GTDB r220 supermatrix:
• 5,869 taxa
• 55% of columns pruned
• Biased taxa: 95.1% → 2.3%
• Runtime: <2h on 4 cores
→ Known clades recovered — without using very complex C60 or CAT models

GTDB r220 case study (led by @kassipan.bsky.social )
Applied WitChi to the archaeal GTDB r220 supermatrix:
• 5,869 taxa
• 55% of columns pruned
• Biased taxa: 95.1% → 2.3%
• Runtime: <2h on 4 cores
→ Known clades recovered — without using very complex C60 or CAT models
With @kassipan.bsky.social and @danieltamarit.bsky.social
🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...

With @kassipan.bsky.social and @danieltamarit.bsky.social

1/ 🧵 Does MAGs contamination affect the placement of Njord as suggested by Zhang et al, 2025? www.nature.com/articles/s41...
Our updated analysis suggests instead... www.biorxiv.org/content/10.1...

1/ 🧵 Does MAGs contamination affect the placement of Njord as suggested by Zhang et al, 2025? www.nature.com/articles/s41...
Our updated analysis suggests instead... www.biorxiv.org/content/10.1...
PhD student position on microbial genome evolution, focusing on the evolutionary principles underlying bacterial genome architecture.
Please repost and share with talented MSc students in #evobio, bioinformatics or related :)
www.uu.nl/en/organisat...
#MEvoSky #MicroSky

PhD student position on microbial genome evolution, focusing on the evolutionary principles underlying bacterial genome architecture.
Please repost and share with talented MSc students in #evobio, bioinformatics or related :)
www.uu.nl/en/organisat...
#MEvoSky #MicroSky
#ArchaeaSky #Asgard #comics #comedy

#ArchaeaSky #Asgard #comics #comedy
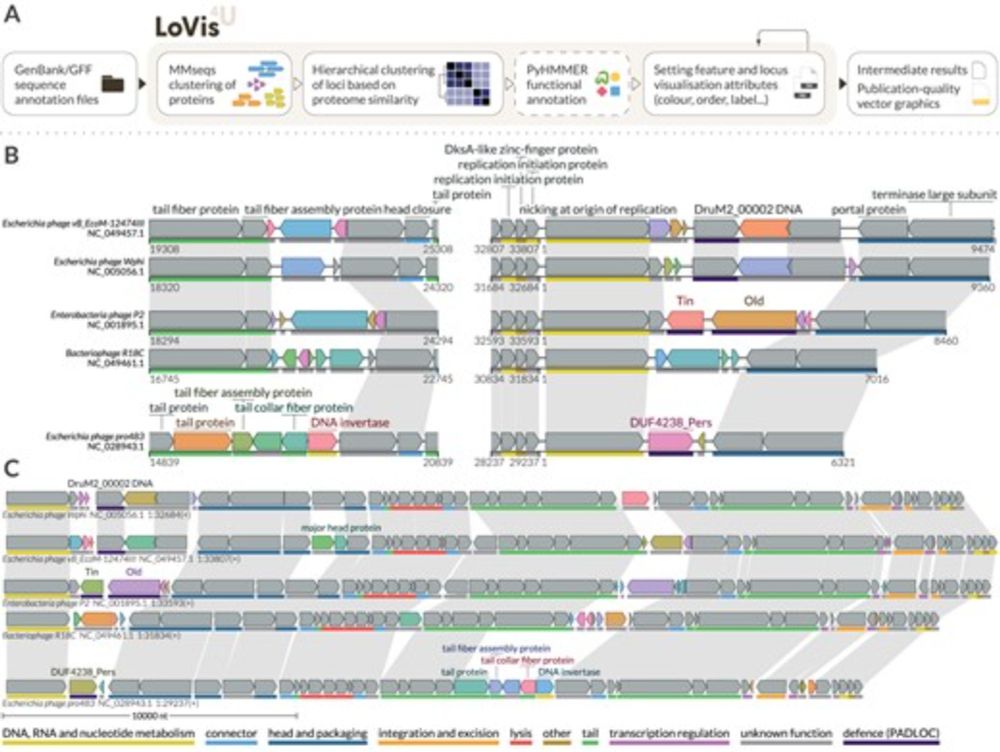


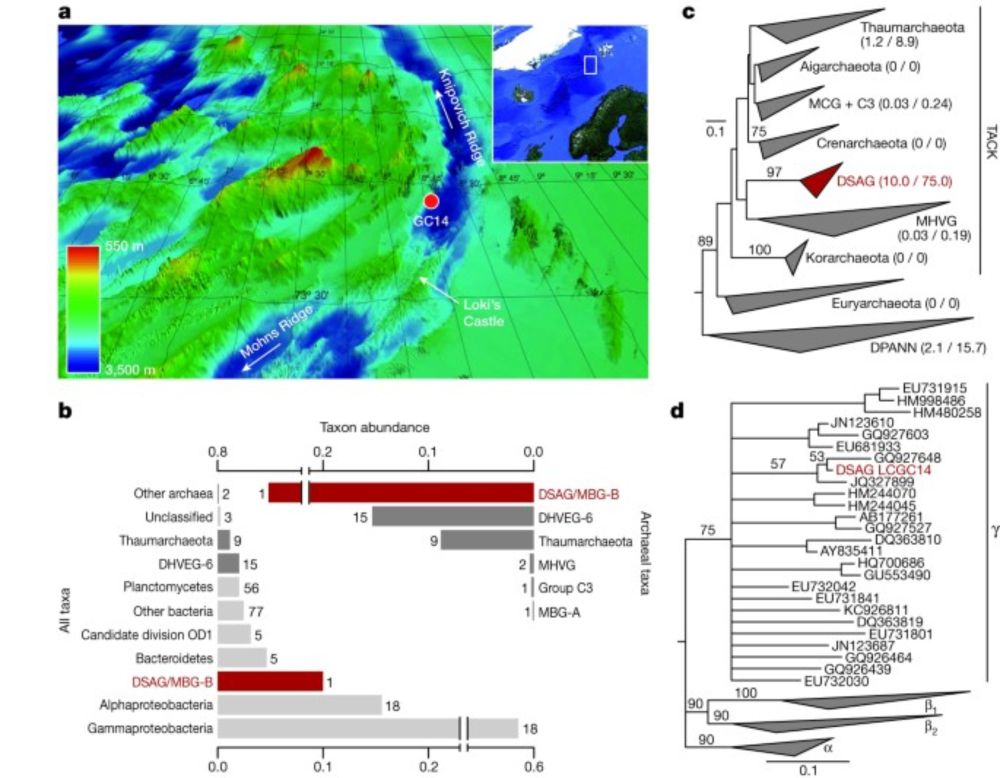
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Could mirror life survive in the wild?
Yes. While mirror life in the wild could have some significant disadvantages (like finding food it can digest), they do not appear to be insurmountable: 🧵

Could mirror life survive in the wild?
Yes. While mirror life in the wild could have some significant disadvantages (like finding food it can digest), they do not appear to be insurmountable: 🧵
You will be part of the EVOLF consortium in which 30 leading biochemists/biophysicists work together to explore if and how we can build autonomous synthetic cells.

You will be part of the EVOLF consortium in which 30 leading biochemists/biophysicists work together to explore if and how we can build autonomous synthetic cells.


