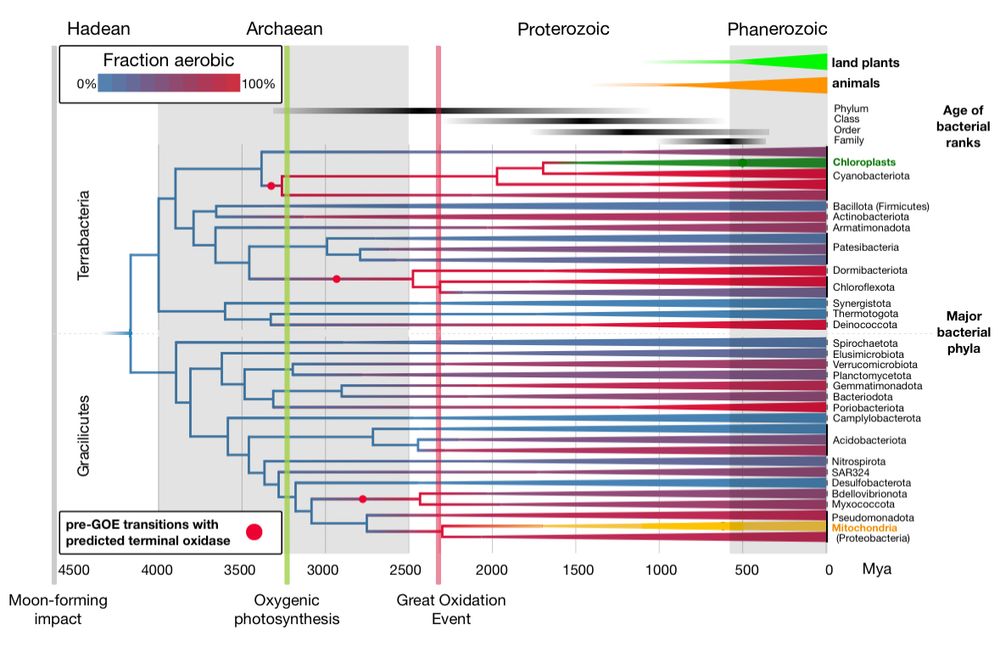
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
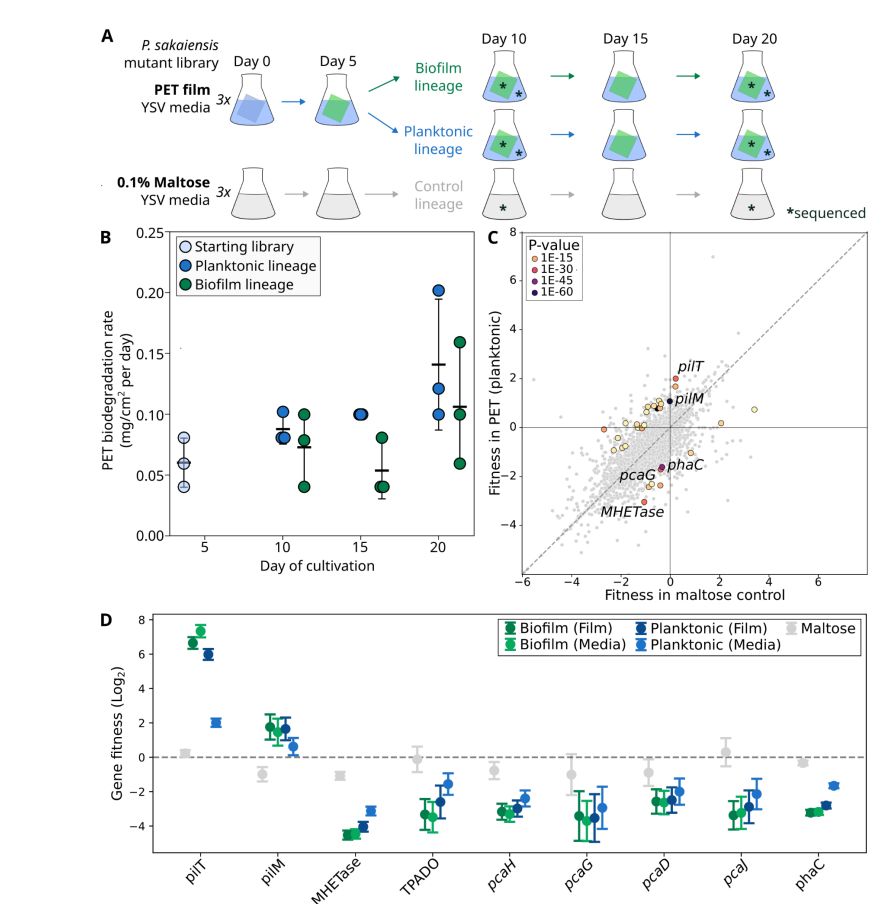
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
After some excellent peer review feedback, a new update of WitChi is taking shape, refining how we detect and prune compositional bias in phylogenomic alignments.
Stay tuned for the next release!
🧙♀️ Can’t model it? Prune it!
github.com/stephkoest/w...

After some excellent peer review feedback, a new update of WitChi is taking shape, refining how we detect and prune compositional bias in phylogenomic alignments.
Stay tuned for the next release!
🧙♀️ Can’t model it? Prune it!
github.com/stephkoest/w...
Applications close November 15!
#phylogenetics #evolution #genomics #ai
evomics.org/apply-worksh...

Applications close November 15!
#phylogenetics #evolution #genomics #ai
evomics.org/apply-worksh...
Stay tuned for two upcoming (PhD student & postdoc) positions to study archaeal genome evolution.
Also huge congrats to my colleague @dorotakawa.bsky.social!
www.uu.nl/en/news/21-v...

Stay tuned for two upcoming (PhD student & postdoc) positions to study archaeal genome evolution.
Also huge congrats to my colleague @dorotakawa.bsky.social!
www.uu.nl/en/news/21-v...
www.archaea.bio/protocols/li...

www.archaea.bio/protocols/li...

Egas @cuwelte.bsky.social et al #microbiology @ribesresearch.bsky.social
www.biorxiv.org/content/10.1...




Egas @cuwelte.bsky.social et al #microbiology @ribesresearch.bsky.social
www.biorxiv.org/content/10.1...
🔗 doi.org/10.1093/molbev/msaf201
#evobio #molbio #archaea

🔗 doi.org/10.1093/molbev/msaf201
#evobio #molbio #archaea
Interested in the evolution of halophily within DPANN archaea we decided to investigate the sister phylum to the Nanohaloarchaeota, GTDB phylum EX4484-52, for which we propose the name Caliditerrarchaeota
www.biorxiv.org/content/10.1...

Interested in the evolution of halophily within DPANN archaea we decided to investigate the sister phylum to the Nanohaloarchaeota, GTDB phylum EX4484-52, for which we propose the name Caliditerrarchaeota
www.biorxiv.org/content/10.1...
Happy to share this one. We investigated the #phylogenetic placement and #genome_evolution of Pangui/Njordarchaea-unique #Asgardarchaea that might have undergone genome reduction...
Happy to share this one. We investigated the #phylogenetic placement and #genome_evolution of Pangui/Njordarchaea-unique #Asgardarchaea that might have undergone genome reduction...

1/ 🧵 Does MAGs contamination affect the placement of Njord as suggested by Zhang et al, 2025? www.nature.com/articles/s41...
Our updated analysis suggests instead... www.biorxiv.org/content/10.1...

1/ 🧵 Does MAGs contamination affect the placement of Njord as suggested by Zhang et al, 2025? www.nature.com/articles/s41...
Our updated analysis suggests instead... www.biorxiv.org/content/10.1...
gtdb.ecogenomic.org
gtdb.ecogenomic.org