
Opinions are my own
he/him

🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...
Please repost
Apply here: www.wur.nl/en/vacancy/p...
Please repost
Apply here: www.wur.nl/en/vacancy/p...
We sat down with @radler92.bsky.social to get more insight into the unique videos from his recent preprint on Promethearchaeota (formerly Asgard archaea).
doi.org/10.1101/2025...
(Videos and info below)

We sat down with @radler92.bsky.social to get more insight into the unique videos from his recent preprint on Promethearchaeota (formerly Asgard archaea).
doi.org/10.1101/2025...
(Videos and info below)
Curious on #niche differentiation of marine #AOA #Nitrosopumilus and #Nitrosopelagicus?
#Urea is (part of) the answer!
Very happy this fun study led by
@mpimarinemicrobio.bsky.social is out:
shorturl.at/wajjh
Big shout out to Joerdis and Hannah for pushing this over the finish line!

Curious on #niche differentiation of marine #AOA #Nitrosopumilus and #Nitrosopelagicus?
#Urea is (part of) the answer!
Very happy this fun study led by
@mpimarinemicrobio.bsky.social is out:
shorturl.at/wajjh
Big shout out to Joerdis and Hannah for pushing this over the finish line!
We tackle a grand challenge in computational chemistry: predicting the structure of crystalline solids directly from their chemical composition.
Paper: arxiv.org/abs/2512.06987
Blog Post: oxtal.github.io
1/8
We tackle a grand challenge in computational chemistry: predicting the structure of crystalline solids directly from their chemical composition.
Paper: arxiv.org/abs/2512.06987
Blog Post: oxtal.github.io
1/8
aithyra.onlyfy.jobs/job/5kc6uod5...
Application deadline: 11.1.2026
#AITHYRA #ProcurementManager

aithyra.onlyfy.jobs/job/5kc6uod5...
Application deadline: 11.1.2026
#AITHYRA #ProcurementManager
aithyra.onlyfy.jobs/job/6vqx92se
Application deadline: 11.1.2026
#AITHYRA #ScientificIllustrator

aithyra.onlyfy.jobs/job/6vqx92se
Application deadline: 11.1.2026
#AITHYRA #ScientificIllustrator
Every day we open a new door onto a different prokaryotic immune system, with extra love for archaeal hosts.
Crafted by the Zink Group within our department :)
Have a wonderful Christmas time! ✨

Every day we open a new door onto a different prokaryotic immune system, with extra love for archaeal hosts.
Crafted by the Zink Group within our department :)
Have a wonderful Christmas time! ✨
Biorxiv.org/content/10.1101/2025.11.30.690169v1

Biorxiv.org/content/10.1101/2025.11.30.690169v1

After some excellent peer review feedback, a new update of WitChi is taking shape, refining how we detect and prune compositional bias in phylogenomic alignments.
Stay tuned for the next release!
🧙♀️ Can’t model it? Prune it!
github.com/stephkoest/w...

After some excellent peer review feedback, a new update of WitChi is taking shape, refining how we detect and prune compositional bias in phylogenomic alignments.
Stay tuned for the next release!
🧙♀️ Can’t model it? Prune it!
github.com/stephkoest/w...

@hohlbeinlab.bsky.social @nicoc-micsynmet.bsky.social @ettema.bsky.social @mib-wur.bsky.social
doi.org/10.1038/s414...

But here I am simulating chromosomes & shuffling motifs.
Watching @loreoliv.bsky.social and the lab turn those predictions into real data was pure magic.✨
Grateful to be part of this team!
But here I am simulating chromosomes & shuffling motifs.
Watching @loreoliv.bsky.social and the lab turn those predictions into real data was pure magic.✨
Grateful to be part of this team!
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
We used comparative genomics on 72,000+ bacterial genomes to uncover the genetic basis of microbial adaptation to multicellular hosts—plants and animals alike.
www.biorxiv.org/content/10.1...

We used comparative genomics on 72,000+ bacterial genomes to uncover the genetic basis of microbial adaptation to multicellular hosts—plants and animals alike.
www.biorxiv.org/content/10.1...
We tested it on the GTDB r220 archaeal supermatrix (5,869 taxa & 10,101 cols) removing 55% of sites in <2h.
The phylogeny showed several interesting groupings with overall improved branch support:
#phylogenetics #ArchaeaSky #MSA #opensource #MEvoSky #MicroSky

We tested it on the GTDB r220 archaeal supermatrix (5,869 taxa & 10,101 cols) removing 55% of sites in <2h.
The phylogeny showed several interesting groupings with overall improved branch support:
#phylogenetics #ArchaeaSky #MSA #opensource #MEvoSky #MicroSky
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...

🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...
Read their groundbreaking study in Current Biology (@currentbiology.bsky.social):
🔗 cemess.univie.ac.at/news/detail-...
Read their groundbreaking study in Current Biology (@currentbiology.bsky.social):
🔗 cemess.univie.ac.at/news/detail-...
Read their groundbreaking study in Current Biology (@currentbiology.bsky.social):
🔗 cemess.univie.ac.at/news/detail-...

or
understanding chromosome organization's deep evolutionary roots
#MicroSky #Archaea #ArchaeaSky #ProtistsOnSky et al.

or
understanding chromosome organization's deep evolutionary roots
#MicroSky #Archaea #ArchaeaSky #ProtistsOnSky et al.
🚨Out Now
Phylogenomic analyses indicate the archaeal superphylum DPANN originated from free-living euryarchaeal-like ancestors, by @bbaker24.bsky.social @deemteam.bsky.social and team
#MicroSky #Archaea 🦠
www.nature.com/articles/s41...
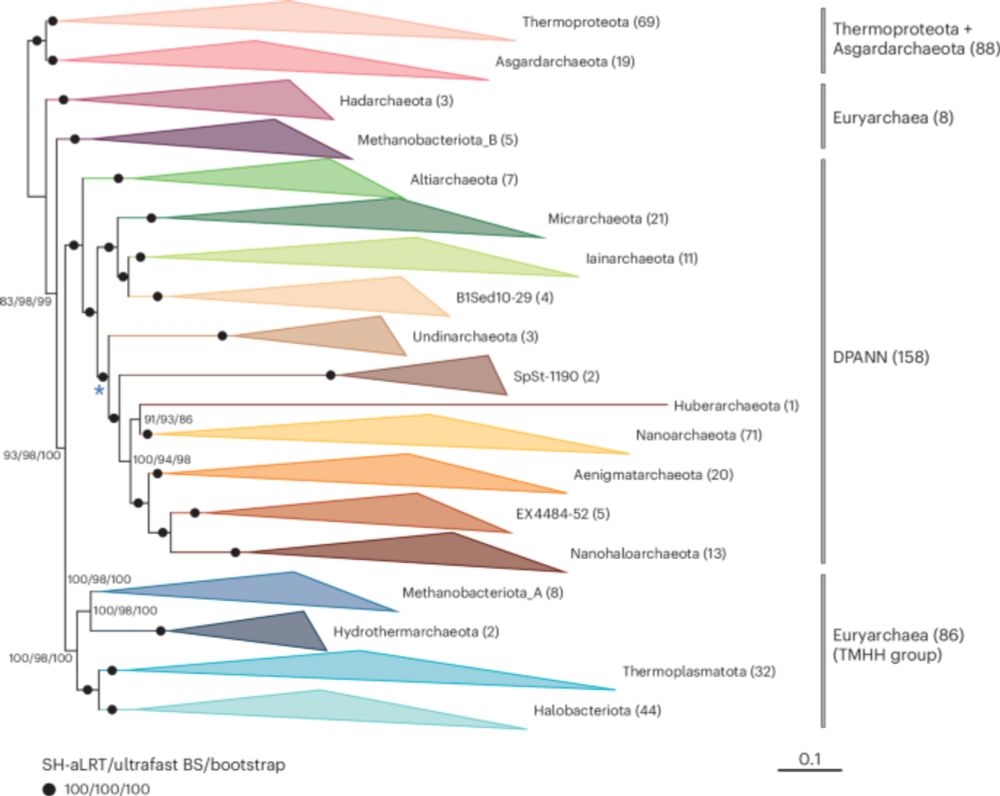
🚨Out Now
Phylogenomic analyses indicate the archaeal superphylum DPANN originated from free-living euryarchaeal-like ancestors, by @bbaker24.bsky.social @deemteam.bsky.social and team
#MicroSky #Archaea 🦠
www.nature.com/articles/s41...



