

#NMRevents #NMRchat #NMR 🧲

#NMRevents #NMRchat #NMR 🧲
erictopol.substack.com/p/our-braini...
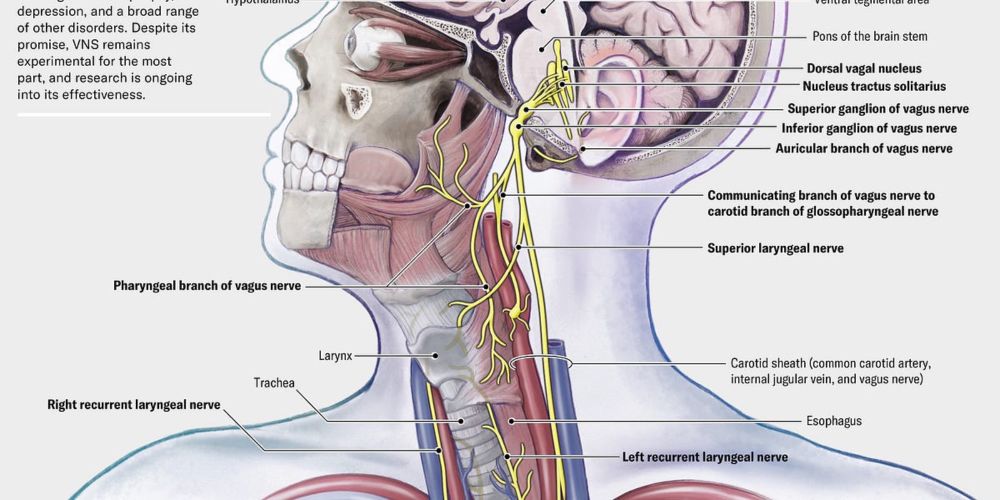
erictopol.substack.com/p/our-braini...
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
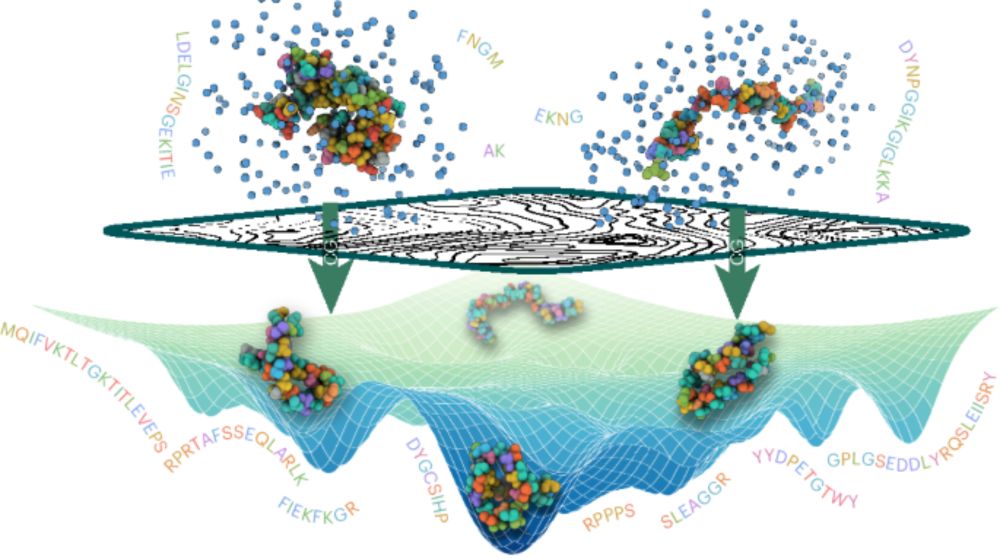
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...



foldingathome.org/2025/06/26/e...
foldingathome.org/2025/06/26/e...




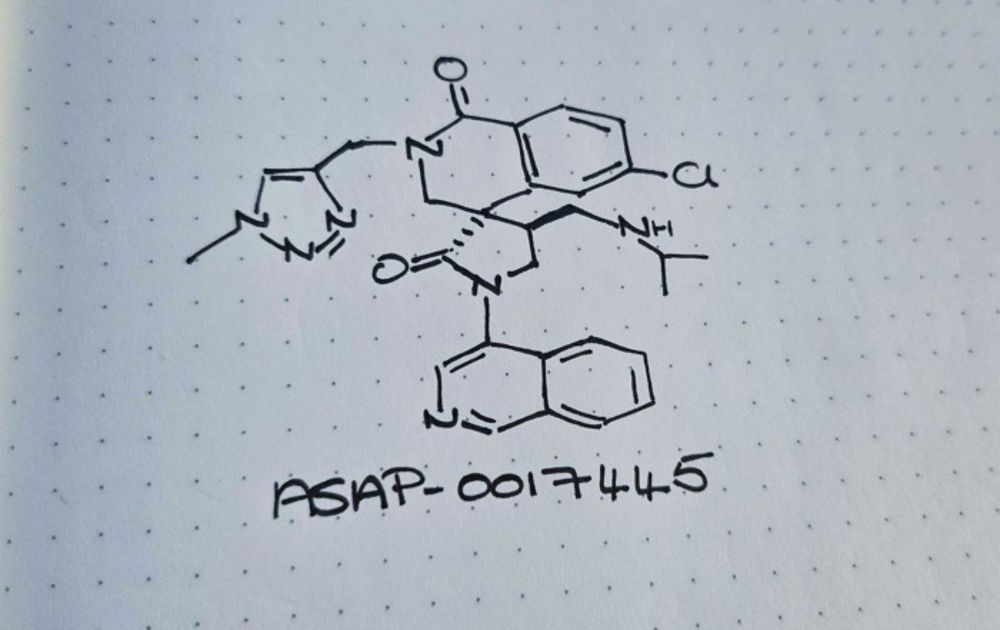

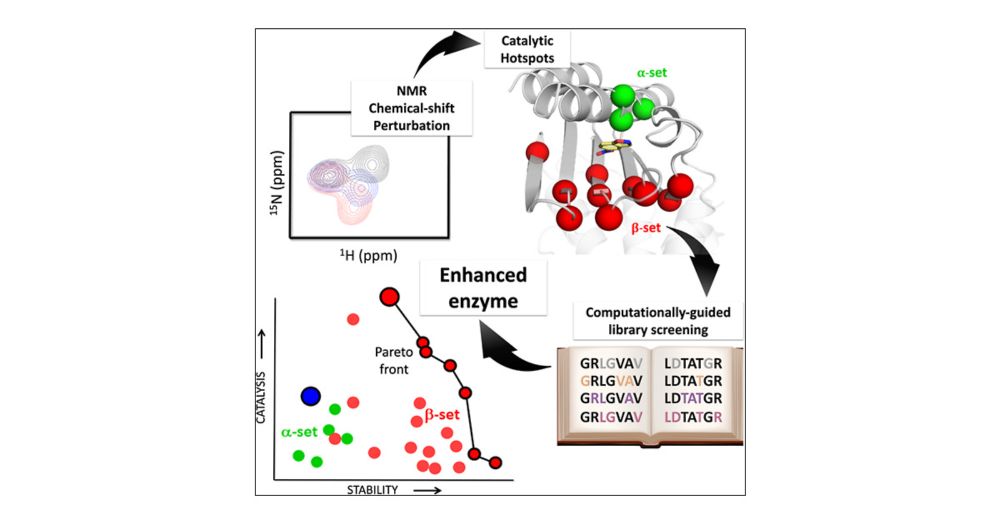

Reference: jamanetwork.com/journals/jam...

Reference: jamanetwork.com/journals/jam...
With @saureli.bsky.social @valeriorizzi.bsky.social and Nicola we used OneOPES to fully converge the free energy landscapes associated with B1AR apo/holo activation.
With @saureli.bsky.social @valeriorizzi.bsky.social and Nicola we used OneOPES to fully converge the free energy landscapes associated with B1AR apo/holo activation.



