Open Science, Proteins and Lipids.
We computed 300 trillion alignments across 1,542 species to map the tree of life. 🧵👇 (1/5)

We computed 300 trillion alignments across 1,542 species to map the tree of life. 🧵👇 (1/5)
Our team has discovered two new bacteriophages that target Serratia spp.—a major cause of hospital‑associated infections and a growing antibiotic‑resistance threat.
📄 Open‑access article in Journal of Applied Microbiology: doi.org/10.1093/jambio/lxag011

Our team has discovered two new bacteriophages that target Serratia spp.—a major cause of hospital‑associated infections and a growing antibiotic‑resistance threat.
📄 Open‑access article in Journal of Applied Microbiology: doi.org/10.1093/jambio/lxag011

www.findaphd.com/phds/project...

www.findaphd.com/phds/project...
📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...

📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...
Very useful as a hypothesis generator, complementing protein interaction prediction from Alphafold.
www.biorxiv.org/content/10.6...
Very useful as a hypothesis generator, complementing protein interaction prediction from Alphafold.
www.biorxiv.org/content/10.6...

We present MemPrO – a toolkit for orienting and building membrane protein systems:
✅ Membrane protein orientation ✅ Mixed membrane building ✅ Double membranes & curvature ✅ Peptidoglycan cell wall support
🔗 github.com/pstansfeld/mempro 📖 pubs.acs.org/doi/10.1021/...

We present MemPrO – a toolkit for orienting and building membrane protein systems:
✅ Membrane protein orientation ✅ Mixed membrane building ✅ Double membranes & curvature ✅ Peptidoglycan cell wall support
🔗 github.com/pstansfeld/mempro 📖 pubs.acs.org/doi/10.1021/...

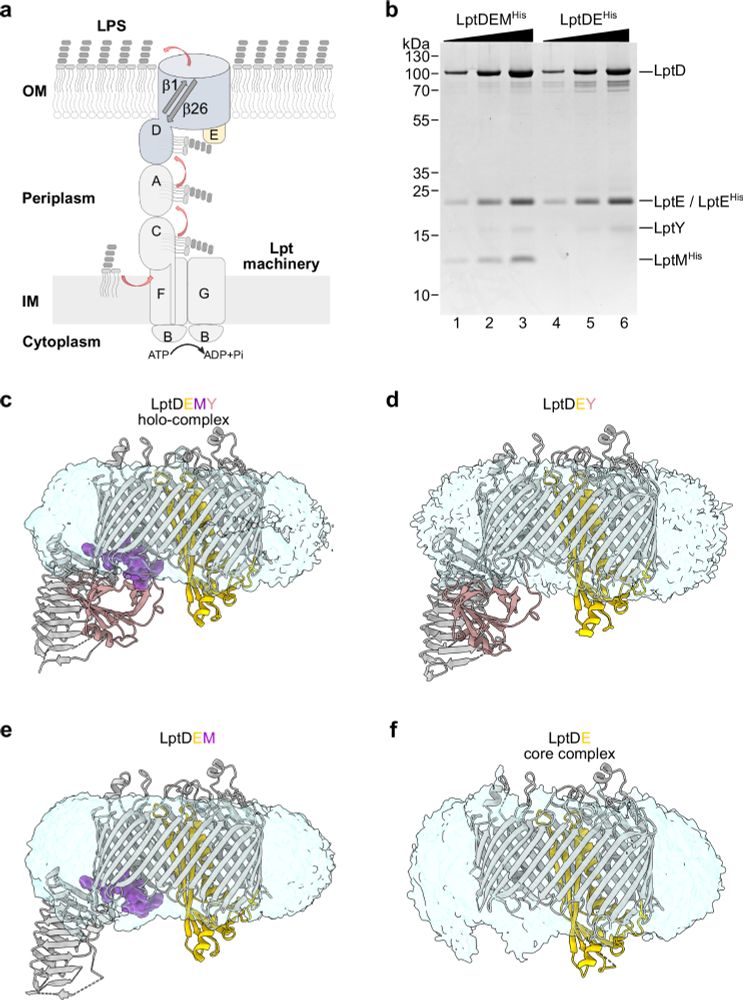

www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...
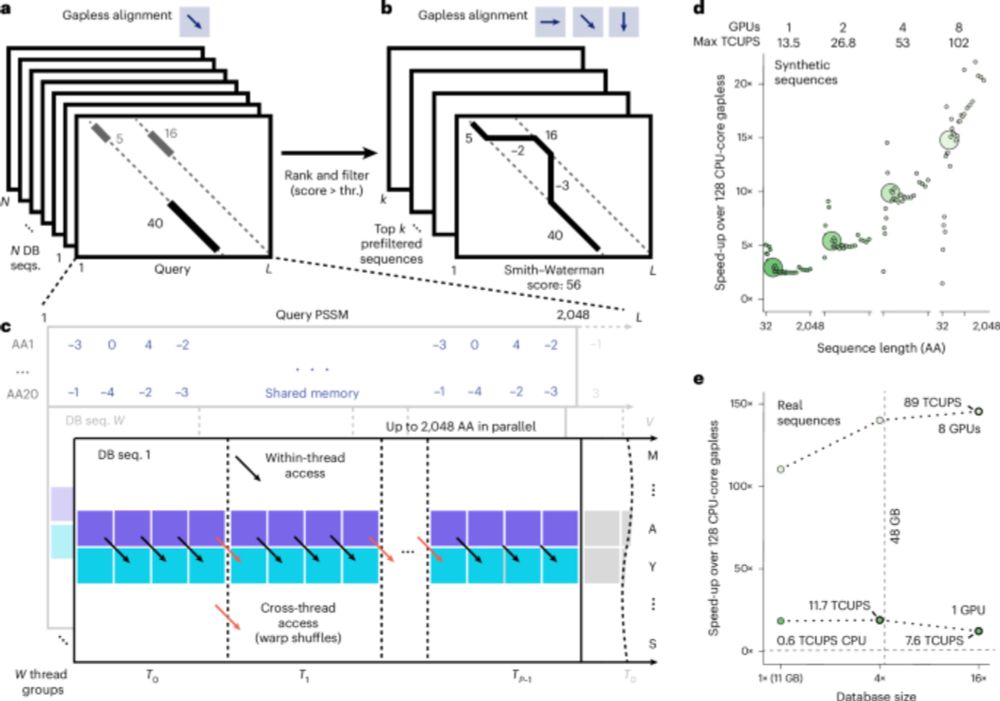
www.nature.com/articles/s41...

We challenge the long-standing view that peptidoglycan alone protects cells from bursting.
Our study shows that the periplasm — enclosed by OM–PG connections — acts as a pressure buffer essential for osmoprotection in Gram-negative bacteria.
📄 www.nature.com/articles/s41...
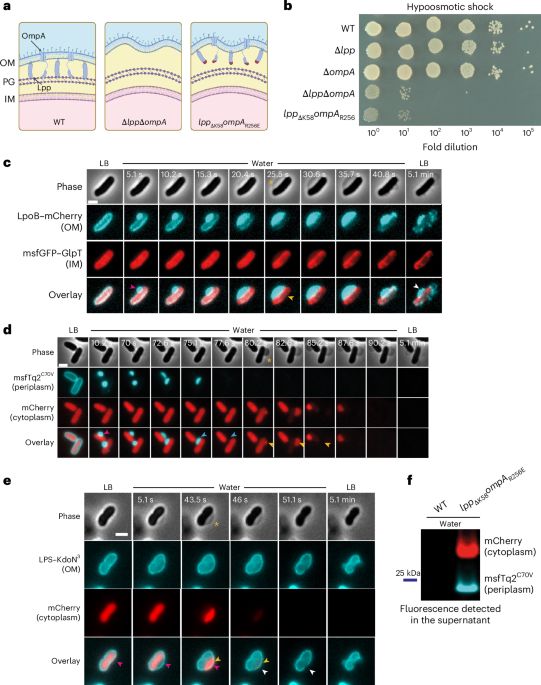
We challenge the long-standing view that peptidoglycan alone protects cells from bursting.
Our study shows that the periplasm — enclosed by OM–PG connections — acts as a pressure buffer essential for osmoprotection in Gram-negative bacteria.
📄 www.nature.com/articles/s41...

In this new paper with colleagues @Bicycle_tx we show a new way to hit old targets: rdcu.be/eoaRI
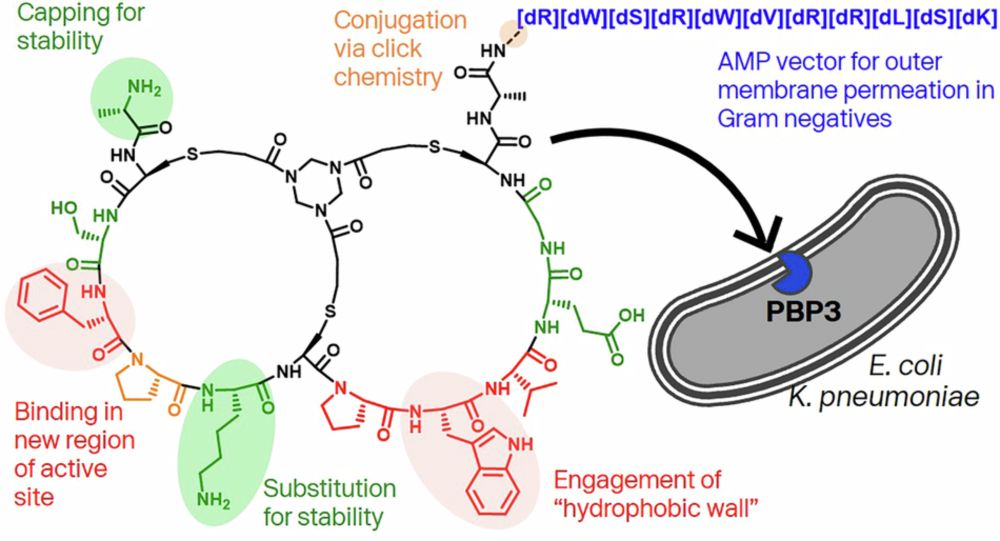
In this new paper with colleagues @Bicycle_tx we show a new way to hit old targets: rdcu.be/eoaRI
👏 French farmers & scientists prove nature-based farming (no #pesticides, no synthetics), the way humans farmed for centuries, still delivers steady yields, stable incomes & boosts biodiversity after 30 years.
#ClimateAction
f24.my/BCO5.BS

👏 French farmers & scientists prove nature-based farming (no #pesticides, no synthetics), the way humans farmed for centuries, still delivers steady yields, stable incomes & boosts biodiversity after 30 years.
#ClimateAction
f24.my/BCO5.BS
www.microbiologyresearch.org/content/jour...

www.microbiologyresearch.org/content/jour...

https://go.nature.com/4iHBBKS

https://go.nature.com/4iHBBKS


