
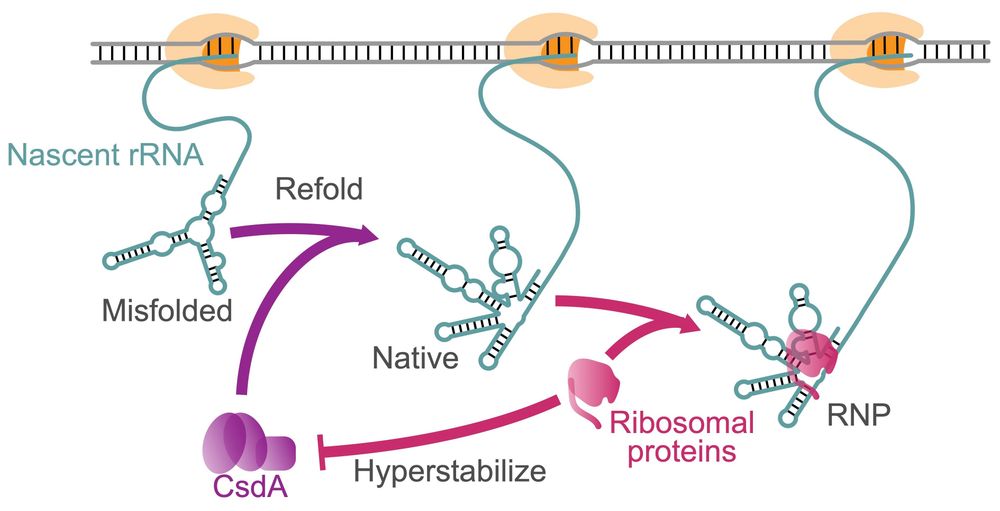
I recently defended my PhD and I'm loved in #RNA structures in vivo 🫶 Very excited to share my two articles at NAR (results of my PhD)!
1📎 academic.oup.com/nar/article/...
2📎 academic.oup.com/nar/article/...
The first #RNAstructure of Ty1 and Ty3 LTR #retrotransposon genomes!

www.nature.com/articles/s41...

www.nature.com/articles/s41...
It shows that the importance of #RNA research in vivo and #RNA genome dynamics of LTR retrotransposons is understood and appreciated by society! #RNAworld ❤️🧬
www.poznan.pl/mim/info/new...

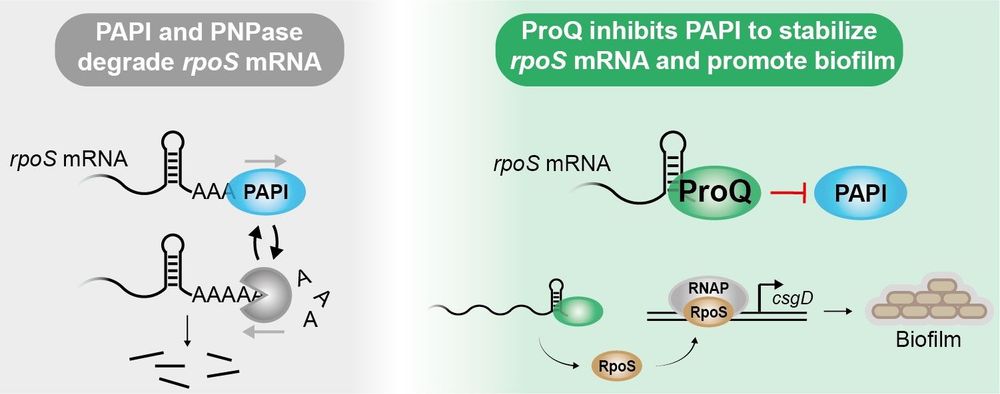
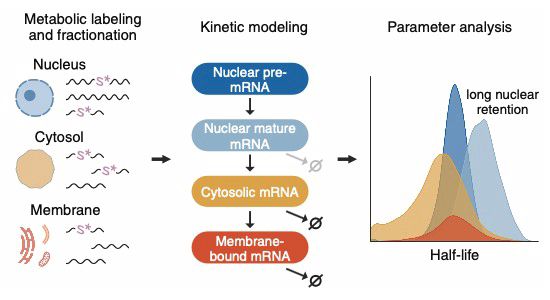
With @karlaneugebauer.bsky.social, @isaac-vock.bsky.social, @mattdsimon.bsky.social
www.cell.com/molecular-ce...

With @karlaneugebauer.bsky.social, @isaac-vock.bsky.social, @mattdsimon.bsky.social
www.cell.com/molecular-ce...
Don’t forget to register for the 2nd Polish RNA Biology Meeting in Poznan, June 24-27! 🧬✨ It’s gonna be an incredible event with an AMAZING lineup of keynote speakers already confirmed! 🎉#RNA
👉 Register here: 2ndrna.web.amu.edu.pl

Don’t forget to register for the 2nd Polish RNA Biology Meeting in Poznan, June 24-27! 🧬✨ It’s gonna be an incredible event with an AMAZING lineup of keynote speakers already confirmed! 🎉#RNA
👉 Register here: 2ndrna.web.amu.edu.pl
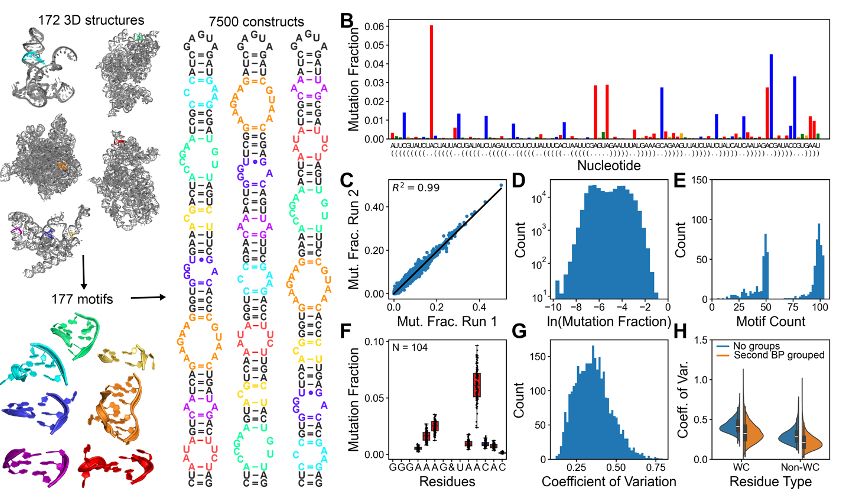
𝐑𝟐𝐃𝐓: a comprehensive platform for visualising RNA secondary structure
Check it out in @narjournal.bsky.social:
academic.oup.com/nar/article/...
Here’s why you should give it a try 🧵👇

𝐑𝟐𝐃𝐓: a comprehensive platform for visualising RNA secondary structure
Check it out in @narjournal.bsky.social:
academic.oup.com/nar/article/...
Here’s why you should give it a try 🧵👇
Cell Reports issue.
😊🤠
Except for insights into RBPs, you will find there also something about circRNAs (of course!) :)
www.cell.com/cell-reports...

Cell Reports issue.
😊🤠
Except for insights into RBPs, you will find there also something about circRNAs (of course!) :)
www.cell.com/cell-reports...
Introducing CaCoFold-R3D by
@aakaran31.bsky.social and myself @rivaselenarivas.bsky.social

And a diploma for my birthday! 🎂

And a diploma for my birthday! 🎂
G4RP.v1 in @natureportfolio.bsky.social: doi.org/10.1038/s414... & nature.com/articles/s41...
& G4RP.v2 now in @cellpressnews.bsky.social: doi.org/10.1016/j.xp...

G4RP.v1 in @natureportfolio.bsky.social: doi.org/10.1038/s414... & nature.com/articles/s41...
& G4RP.v2 now in @cellpressnews.bsky.social: doi.org/10.1016/j.xp...
www.nature.com/articles/s42...

www.nature.com/articles/s42...
congrats to Dr. Lu Xiao!
doi: 10.1016/j.chembiol.2024.09.009

congrats to Dr. Lu Xiao!
doi: 10.1016/j.chembiol.2024.09.009

The NIH BioArt Source is an awesome library of *free* professionally drawn illustrations for scientific presentations or figures. Downloadable in HD. Thank you NIH for this invaluable tool 🙏!
Check it out 👇
bioart.niaid.nih.gov

The NIH BioArt Source is an awesome library of *free* professionally drawn illustrations for scientific presentations or figures. Downloadable in HD. Thank you NIH for this invaluable tool 🙏!
Check it out 👇
bioart.niaid.nih.gov
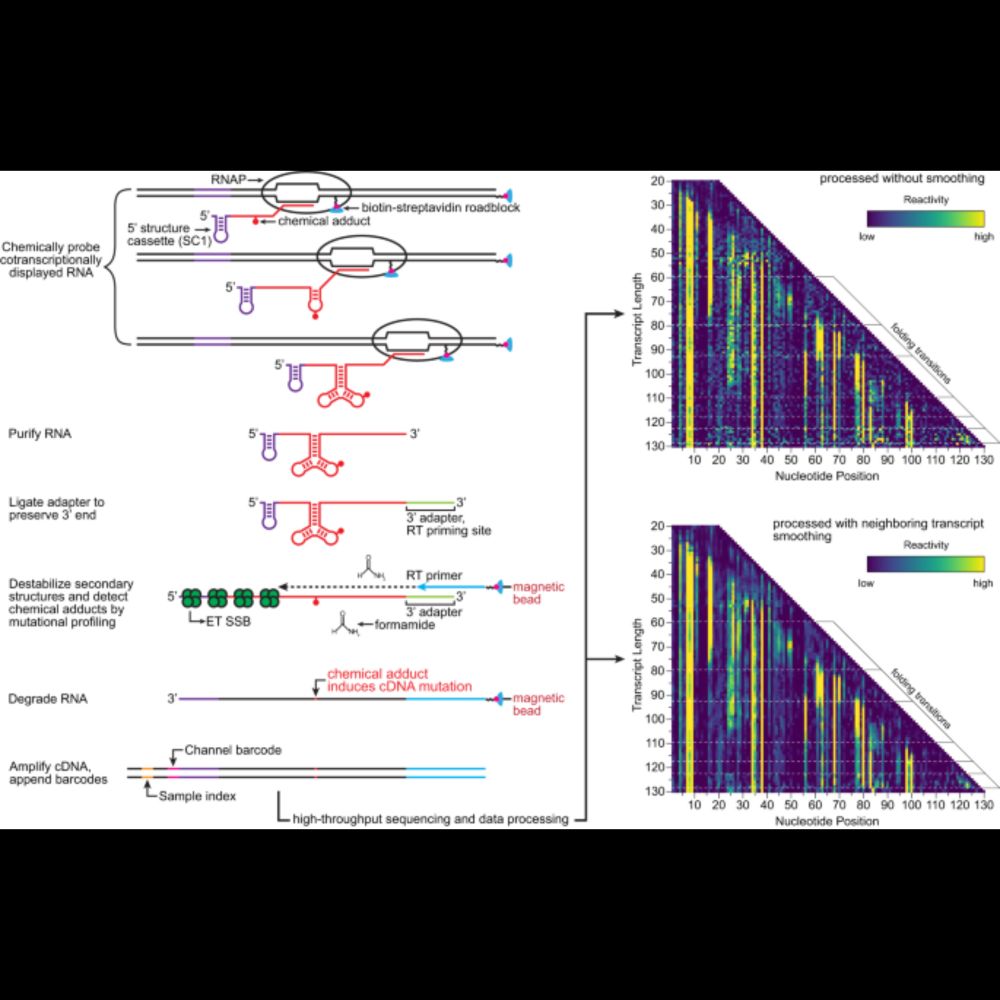
Such a great RNA community being built here! Thought I’d introduce our wonderful RNA structure drawing app RNAcanvas. 100’s of daily users all over the world
rna2drawer.app
RNAcanvas: interactive drawing and exploration of nucleic acid structures academic.oup.com/nar/article/...

Such a great RNA community being built here! Thought I’d introduce our wonderful RNA structure drawing app RNAcanvas. 100’s of daily users all over the world
rna2drawer.app
RNAcanvas: interactive drawing and exploration of nucleic acid structures academic.oup.com/nar/article/...
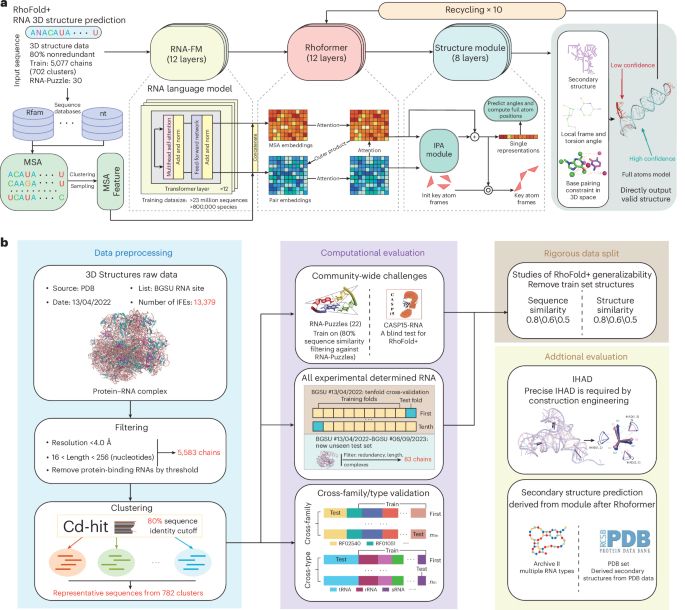
I want to have it here!
This cover is an extraordinary honor for me and shows me that it is worth trying even if the chances are small!
Related study here 👇
academic.oup.com/nar/article/...

I want to have it here!
This cover is an extraordinary honor for me and shows me that it is worth trying even if the chances are small!
Related study here 👇
academic.oup.com/nar/article/...
I recently defended my PhD and I'm loved in #RNA structures in vivo 🫶 Very excited to share my two articles at NAR (results of my PhD)!
1📎 academic.oup.com/nar/article/...
2📎 academic.oup.com/nar/article/...
The first #RNAstructure of Ty1 and Ty3 LTR #retrotransposon genomes!

I recently defended my PhD and I'm loved in #RNA structures in vivo 🫶 Very excited to share my two articles at NAR (results of my PhD)!
1📎 academic.oup.com/nar/article/...
2📎 academic.oup.com/nar/article/...
The first #RNAstructure of Ty1 and Ty3 LTR #retrotransposon genomes!


