I don’t know his much detail is in the open of ONT’s Bonito basecaller
I don’t know his much detail is in the open of ONT’s Bonito basecaller
github.com/google/deepv...
- 1.6x time reduction (40% increase ??)
- Pangenome integration
- New model and improvements over previously model
Excited to try this new update 🧬💻
#bioinformatics #wgs #ngs #variantcalling

github.com/google/deepv...
- 1.6x time reduction (40% increase ??)
- Pangenome integration
- New model and improvements over previously model
Excited to try this new update 🧬💻
#bioinformatics #wgs #ngs #variantcalling

What reference genome should you use?
Sounds easy. It’s not.
GRCh37? GRCh38? hs37d5?
Have you heard of T2T or the new pan-genome-aware DeepVariant?
This matters more than you think.
www.biorxiv.org/content/10....
What reference genome should you use?
Sounds easy. It’s not.
GRCh37? GRCh38? hs37d5?
Have you heard of T2T or the new pan-genome-aware DeepVariant?
This matters more than you think.
www.biorxiv.org/content/10....
What reference genome should you use?
Sounds easy. It’s not.
GRCh37? GRCh38? hs37d5?
Have you heard of T2T or the new pan-genome-aware DeepVariant?
This matters more than you think.
www.biorxiv.org/content/10....
What reference genome should you use?
Sounds easy. It’s not.
GRCh37? GRCh38? hs37d5?
Have you heard of T2T or the new pan-genome-aware DeepVariant?
This matters more than you think.
www.biorxiv.org/content/10....
But those are specialized models, not these LLMs that are taking over everywhere else
But those are specialized models, not these LLMs that are taking over everywhere else

Illumina+Nanopore data & DeepVariant enhance accurate, cost-effective germline variant calling, improving diagnostics.




Illumina+Nanopore data & DeepVariant enhance accurate, cost-effective germline variant calling, improving diagnostics.
bsky.app/profile/nano...

bsky.app/profile/nano...
You get the idea
You get the idea

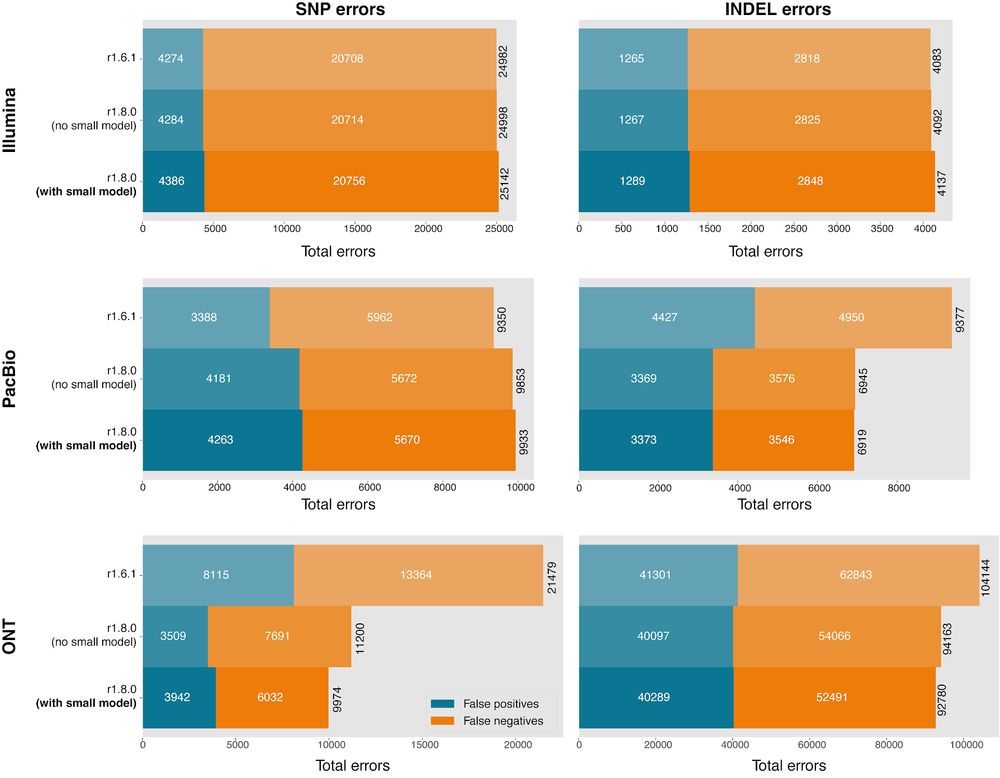
github.com/google/deepv...
Shows a step by step process, with Docker images for how to map to a Pangenome reference w/ vg and calls w/ DeepVariant. Final calls are more accurate and in GRCh38 coordinates. Thanks to the UCSC team for co-development
github.com/google/deepv...
Shows a step by step process, with Docker images for how to map to a Pangenome reference w/ vg and calls w/ DeepVariant. Final calls are more accurate and in GRCh38 coordinates. Thanks to the UCSC team for co-development
Support for haploid regions, chrX/Y.
Workflow for Pangenome FASTQ-to-VCF.
Major DeepTrio improvements for de novo variants.
Models for CompleteGenomics T7, G400
Add NovaSeqX to training data
Release by Kishwar Shafin
github.com/google/deepv...

Support for haploid regions, chrX/Y.
Workflow for Pangenome FASTQ-to-VCF.
Major DeepTrio improvements for de novo variants.
Models for CompleteGenomics T7, G400
Add NovaSeqX to training data
Release by Kishwar Shafin
github.com/google/deepv...
While it's nice to see comparisons, why compare an (at the time) 2 year old GATK against a 5 year old bcftools?
Since then both have come on a lot. It'd be interesting to see new independent comparisons. (Neither can hold up to deepvariant now.)

While it's nice to see comparisons, why compare an (at the time) 2 year old GATK against a 5 year old bcftools?
Since then both have come on a lot. It'd be interesting to see new independent comparisons. (Neither can hold up to deepvariant now.)

