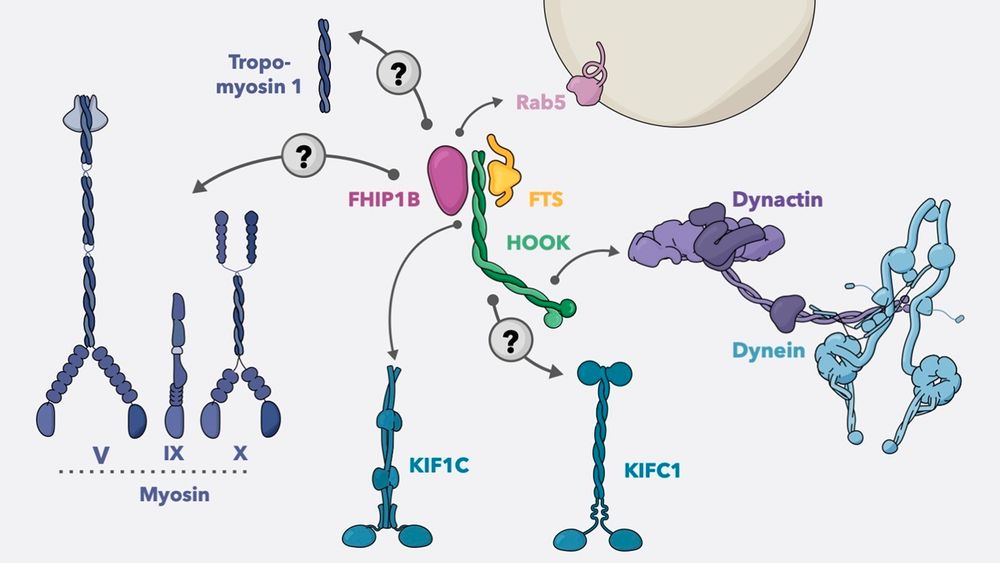
Cryo-ET🔬reveals a large new domain on the small subunit, built from multiple extensions in conserved ribosomal proteins.
bioRxiv 📖: shorturl.at/q44tG
This suggests greater chlororibosome diversity than expected!
1/n 🧵
We found that the proteins Nono and SFPQ assemble into long filaments. Curious to know whether these filaments also exist in paraspeckles.
Very enjoyable collaboration with Dietmar Geiger at @uni-wuerzburg.de . We used #cryoEM for structure determination rdcu.be/e06t7 .

We found that the proteins Nono and SFPQ assemble into long filaments. Curious to know whether these filaments also exist in paraspeckles.
Very enjoyable collaboration with Dietmar Geiger at @uni-wuerzburg.de . We used #cryoEM for structure determination rdcu.be/e06t7 .
Bonus: lots of cryoEM ❄️🔬 references to dig into...
Bonus: lots of cryoEM ❄️🔬 references to dig into...
8:30am-12:30pm, Sat Feb 15, Room 515B
See detailed program at the link below. We have some superb science lined up, so please join us for what I hope will be a fantastic morning of science!

I finally read: Henderson R (1995) The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Quarterly Reviews of Biophysics 28: 171–193 […]
I finally read: Henderson R (1995) The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Quarterly Reviews of Biophysics 28: 171–193 […]
It is an ambitious course. A few lectures about the theory, then mostly practicals all the way […]
It is an ambitious course. A few lectures about the theory, then mostly practicals all the way […]
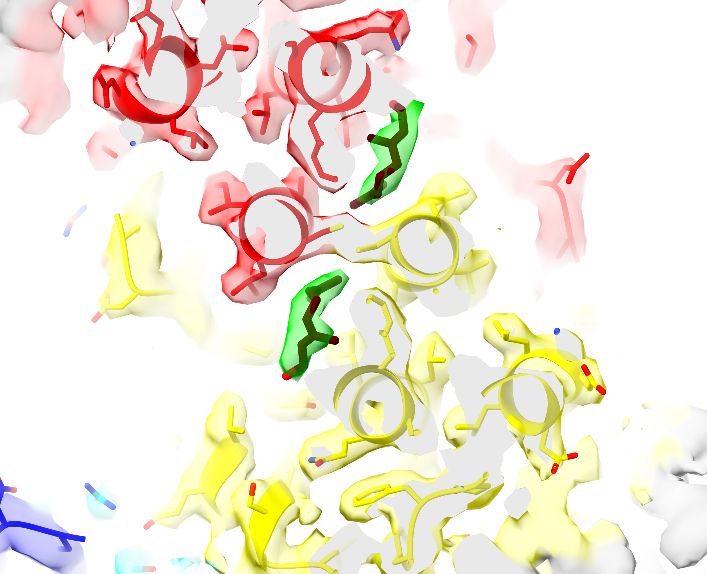
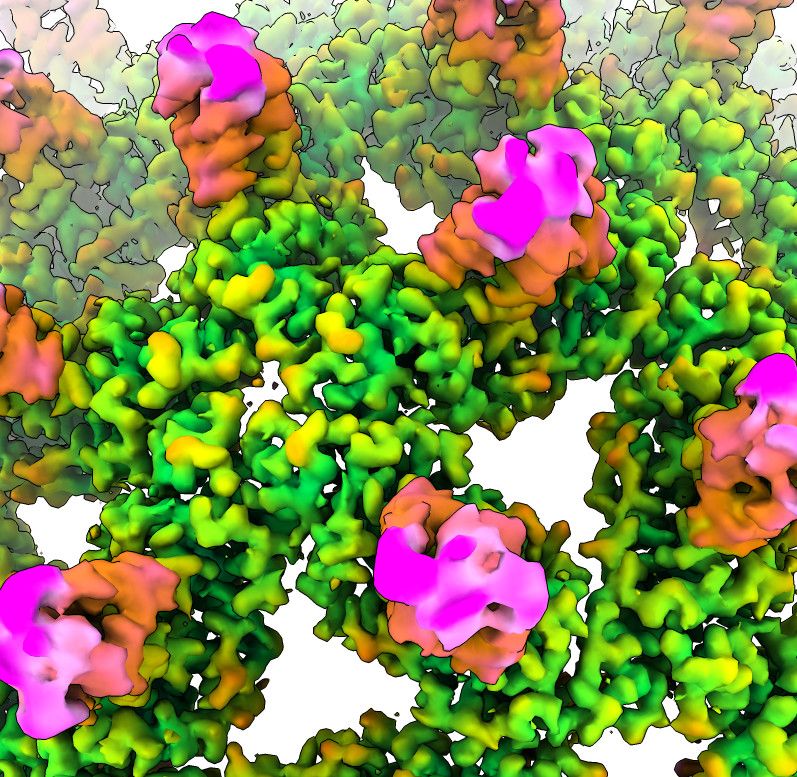


by @jonmarkert.bsky.social @lucas.farnunglab.com et al. at @harvardcellbio.bsky.social #cryoEM
#chromatin #transcription #cryoEM #Pol_II🧪🧬❄️🔬
www.science.org/doi/10.1126/...
by @jonmarkert.bsky.social @lucas.farnunglab.com et al. at @harvardcellbio.bsky.social #cryoEM
#chromatin #transcription #cryoEM #Pol_II🧪🧬❄️🔬
www.science.org/doi/10.1126/...
I recently read: Marques MA, Purdy MD & Yeager M (2019) CryoEM maps are full of potential. Current Opinion in Structural Biology 58: 214–223 https://doi.org10.1016/j.sbi.2019.04.006
It's […]
I recently read: Marques MA, Purdy MD & Yeager M (2019) CryoEM maps are full of potential. Current Opinion in Structural Biology 58: 214–223 https://doi.org10.1016/j.sbi.2019.04.006
It's […]

Do you like it short as “#CryoEM”?
#cryoEM|cryoEM|cryo-EM|#cryoet|❄️🔬|#teamtomo|#insitu|#cryo-ET|cryoET|cryo-ET|#cryofib|cryofib|cryo-fibsem
Do you like it short as “#CryoEM”?
#cryoEM|cryoEM|cryo-EM|#cryoet|❄️🔬|#teamtomo|#insitu|#cryo-ET|cryoET|cryo-ET|#cryofib|cryofib|cryo-fibsem




bsky.app/profile/did:...
#CryoEM ❄️ 🔬
#Science 🧬 🦠
#Education 📚
Huge shoutout to our SPoCs for making this a grand success! 👏
Marzia Miletto
Marcelo Alexandre de Farias
Omar Davulcu
#CryoEM ❄️ 🔬
#Science 🧬 🦠
#Education 📚
Huge shoutout to our SPoCs for making this a grand success! 👏
Marzia Miletto
Marcelo Alexandre de Farias
Omar Davulcu
The course will be for PhD student, postdoc, staff scientist or young group leaders.
If you think it will benefit you go to the website for more info: astbury.leeds.ac.uk/facilities/w...
Sharing will be appreciated :-)