
Postdoc @Cambridge ➡️ERC Pepinsure

---
#proteomics #prot-paper

---
#proteomics #prot-paper

doi.org/10.1016/j.ja...

---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
Apply by April 13, 2025!
Link: employment.ku.dk/staff?show=1...
#LabJobs #BiomolecularScience #AncientDNA
👇

Apply by April 13, 2025!
Link: employment.ku.dk/staff?show=1...
#LabJobs #BiomolecularScience #AncientDNA
👇
"InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments"
🔗 www.nature.com/articles/s42...
👇 Thread 🧵
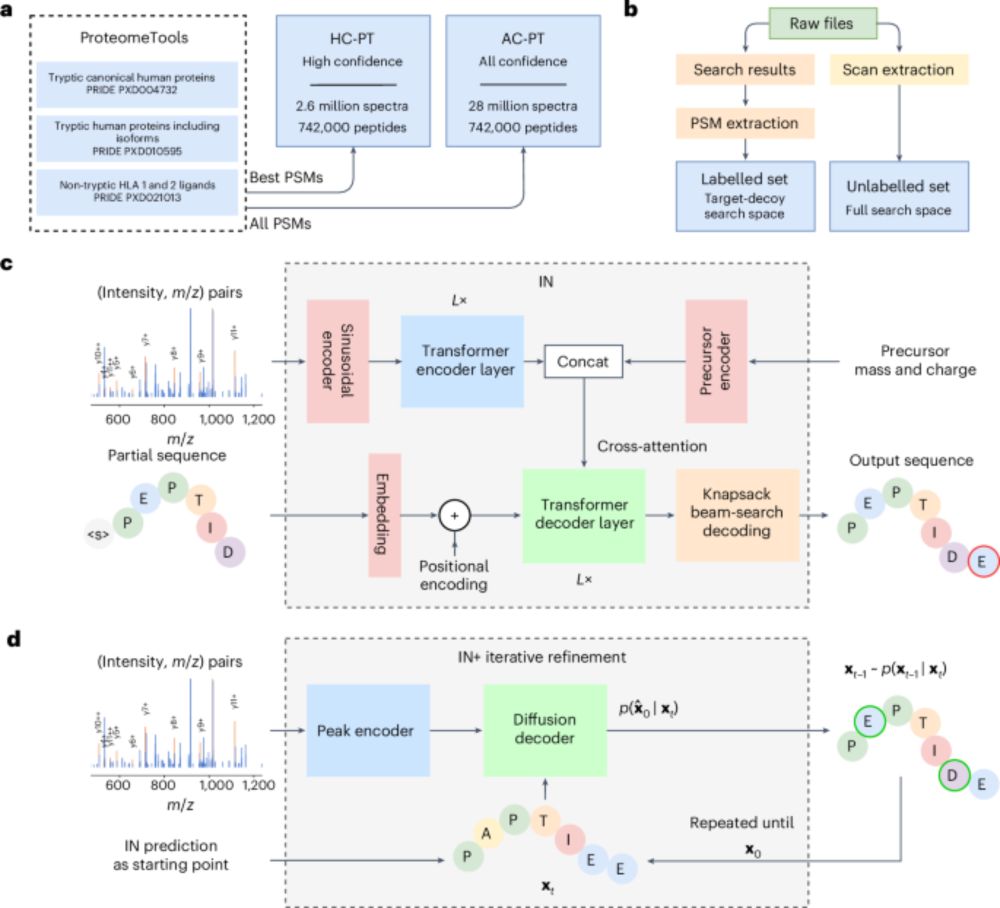
"InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments"
🔗 www.nature.com/articles/s42...
👇 Thread 🧵
Some financial assistance is available - see the course website.
🧪🧶🧬🖥️
www.biorxiv.org/content/10.1...

It is a minor update, main highlights are:
- Built-in support for Thermo .raw (Windows + Linux)
- New QC metrics in PDF report & visualising trends across the experiment

It is a minor update, main highlights are:
- Built-in support for Thermo .raw (Windows + Linux)
- New QC metrics in PDF report & visualising trends across the experiment
github.com/smith-chem-w...

github.com/smith-chem-w...
Feel free to create your own!
#rstats #dataviz #phd

Feel free to create your own!
#rstats #dataviz #phd

pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...
Github: github.com/BAMeScience/...
Paper: www.nature.com/articles/s41...
Many thanks to everyone involved 🙌 #MachineLearning #MassSpec #Metabolomics #FIORA
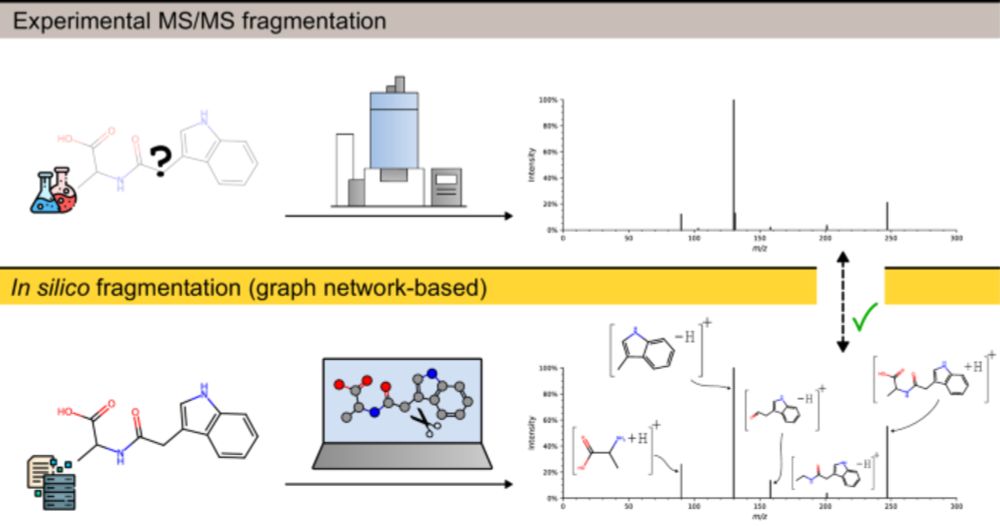
Github: github.com/BAMeScience/...
Paper: www.nature.com/articles/s41...
Many thanks to everyone involved 🙌 #MachineLearning #MassSpec #Metabolomics #FIORA
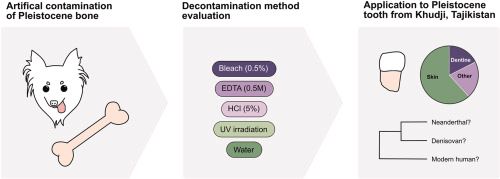
While promising, paleoproteomics requires exceptional methodology to distinguish genuine ancient proteins from contamination. Let's wait for peer review and independent verification. #paleoproteomics #scienceskepticism 🙂
While promising, paleoproteomics requires exceptional methodology to distinguish genuine ancient proteins from contamination. Let's wait for peer review and independent verification. #paleoproteomics #scienceskepticism 🙂
A) Wait for the clearance deals in the coming 6 weeks and take advantage of the deals
B) Wait until after ASMS and then update the order
A) Wait for the clearance deals in the coming 6 weeks and take advantage of the deals
B) Wait until after ASMS and then update the order
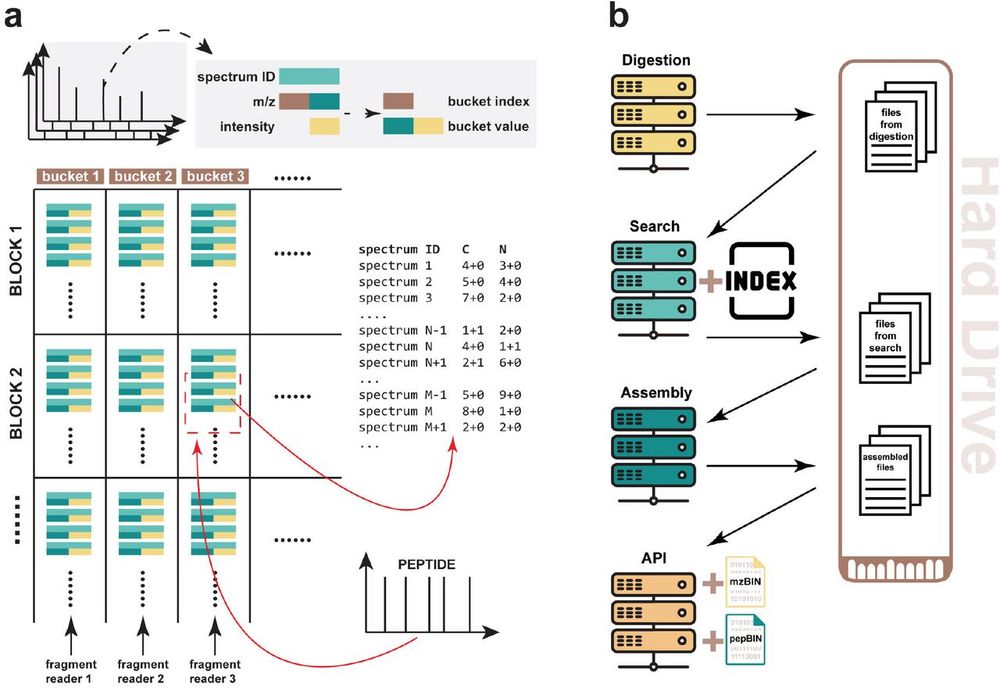


www.nature.com/articles/s41...
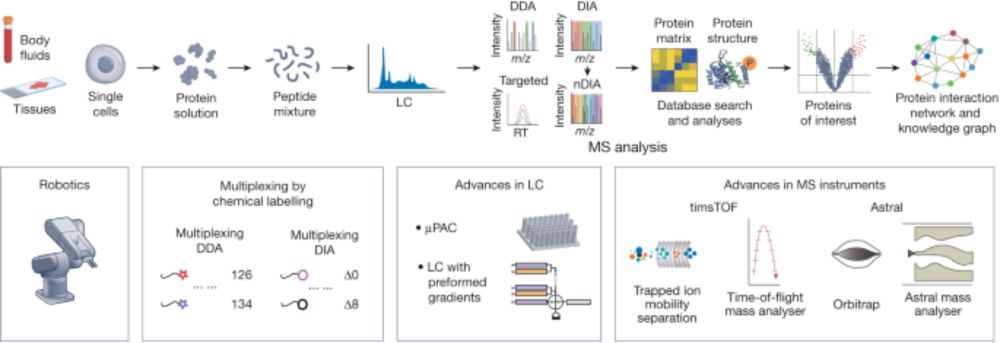
www.nature.com/articles/s41...
github.com/klaudiosinan...
github.com/LazoCoder/Po...


github.com/klaudiosinan...
github.com/LazoCoder/Po...
What was previously reported to have little global impact now is reported to explain up to half of the variation in how protein levels are controlled. Protein degradation matters!
shorturl.at/5wPiS

What was previously reported to have little global impact now is reported to explain up to half of the variation in how protein levels are controlled. Protein degradation matters!
shorturl.at/5wPiS

