
https://andrew-leduc.github.io/
Studying how variation in protein half-life
leads to variation in protein levels
One sphere is black (1 ppm).
Finding the black sphere is comparable to detecting a protein present at ~ 6,000 copies in the proteome of a human cell.
Quantifying the protein requires analyzing multiple jars.

One sphere is black (1 ppm).
Finding the black sphere is comparable to detecting a protein present at ~ 6,000 copies in the proteome of a human cell.
Quantifying the protein requires analyzing multiple jars.
Germinal produces functional nanobodies in just dozens of tests, making custom antibody design more accessible than ever before.

The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules ⬇️
www.biorxiv.org/content/10.1...

The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules ⬇️
www.biorxiv.org/content/10.1...

The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules ⬇️
www.biorxiv.org/content/10.1...
The results revealed 𝐜𝐨𝐦𝐩𝐥𝐞𝐱 regulation & 𝐬𝐢𝐦𝐩𝐥𝐞 organizing principles:
www.biorxiv.org/content/10.1...
🧵

The results revealed 𝐜𝐨𝐦𝐩𝐥𝐞𝐱 regulation & 𝐬𝐢𝐦𝐩𝐥𝐞 organizing principles:
www.biorxiv.org/content/10.1...
🧵
Call to action from the community to achive this ambitious goal!
Call to action from the community to achive this ambitious goal!
Announced in @cellcellpress.bsky.social, the competition is hosted by Arc Institute and sponsored by NVIDIA, @10xgenomics.bsky.social and Ultima Genomics.

They are essentially entirely unquantified by 10x sample preparation, probably because cell lysis is not sufficiently strong.
They are essentially entirely unquantified by 10x sample preparation, probably because cell lysis is not sufficiently strong.
There are some differences in the information they give and would be interesting to explore.
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...
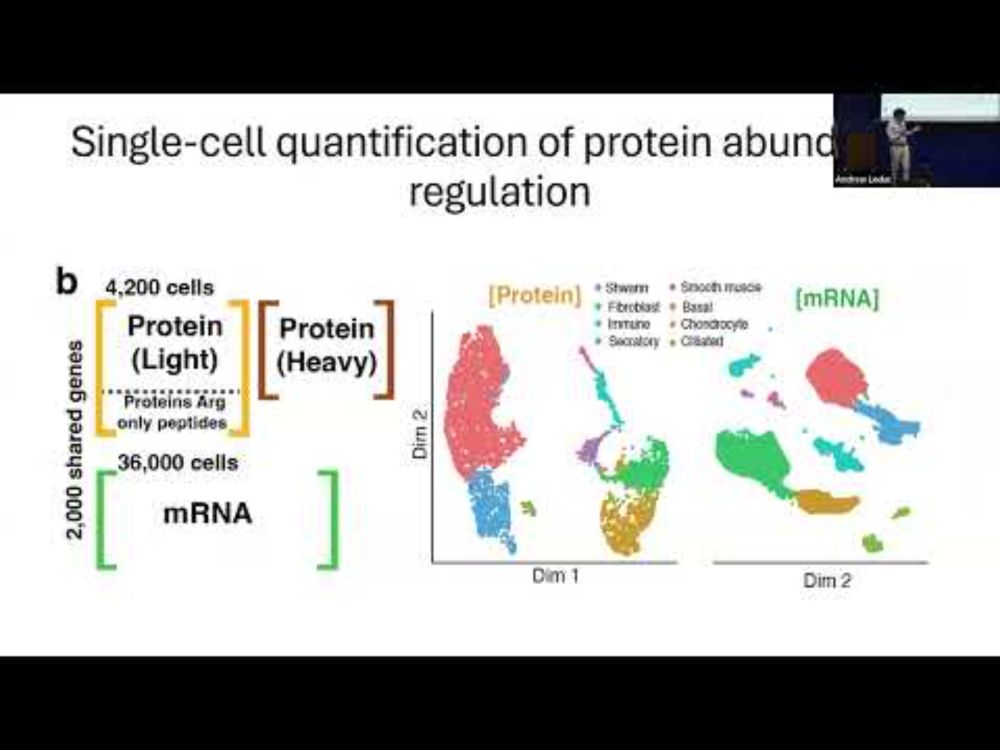
There are some differences in the information they give and would be interesting to explore.
1/n
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...

1/n
Our project focuses on better understanding the regulation of protein abundance by measuring transcription, translation, and protein clearance in single cells.
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...

Our project focuses on better understanding the regulation of protein abundance by measuring transcription, translation, and protein clearance in single cells.
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...

𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...
Wow cells do so much stuff
Wow cells do so much stuff
We have seen them in all tissues that we have analyzed, and @andrewleduc.bsky.social has even more compelling examples from mouse trachea ... soon to be published!
We have seen them in all tissues that we have analyzed, and @andrewleduc.bsky.social has even more compelling examples from mouse trachea ... soon to be published!


We find large & consistent decrease in the abundance and stoichiometry of 20S proteasome subunits.
⬛️ It leads to abnormal accumulation of 20S substrate proteins.
🧵
biorxiv.org/content/10.1...

That’s millions of protein data points per day.
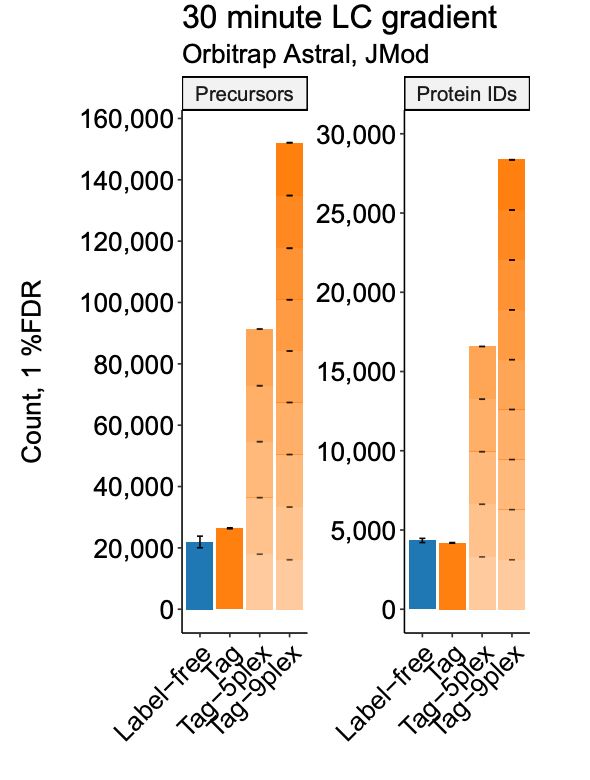
That’s millions of protein data points per day.

That’s millions of protein data points per day.
timePlex (multiplexing with time-offsets): www.biorxiv.org/content/10.1...
JMod (search software for joint-modeling): www.biorxiv.org/content/10.1...
PSMtag (novel mass tag for 9-plexDIA): www.biorxiv.org/content/10.1...
It enables:
-Label-free multiplexing
-Combinatorial multiplexing with plexDIA
Using combined 9-plexDIA and 3-timePlex we demonstrate 27-plex DIA 🚀
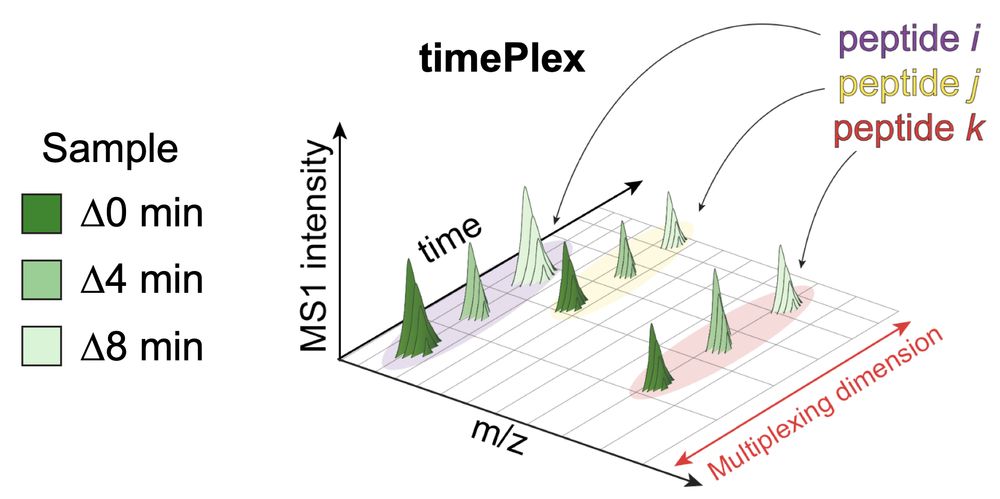
It enables:
-Label-free multiplexing
-Combinatorial multiplexing with plexDIA
Using combined 9-plexDIA and 3-timePlex we demonstrate 27-plex DIA 🚀

