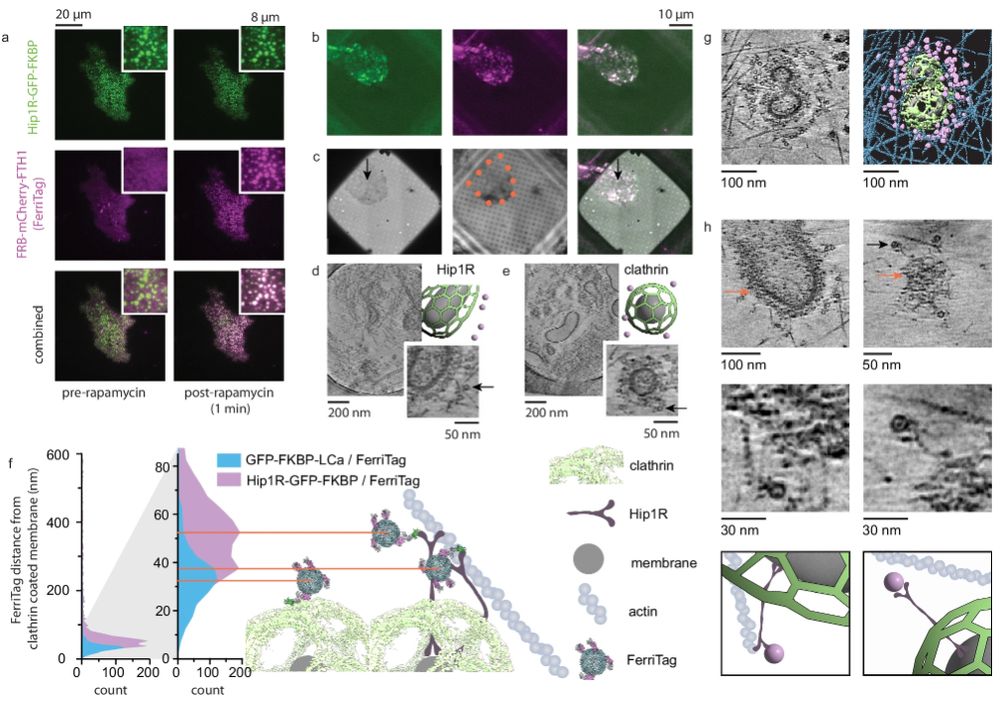
I like to make open source cryoET things.
Learn more about Ultrack, a powerful tool for fast, accurate and scalable cell tracking: biohub.org/life-science... 🧪
www.nature.com/nmeth/volume...
On the cover, cell tracking in a developing zebrafish embryo visualized using Ultrack. Cover by Alexandre Dizieux, Jordão Bragantini, Loic Royer, Merlin Lange, CZ Biohub. Paper here: www.nature.com/articles/s41...

Learn more about Ultrack, a powerful tool for fast, accurate and scalable cell tracking: biohub.org/life-science... 🧪

Kaggle cryo-ET picking: Over-pick tol, aug, & annot key.




Kaggle cryo-ET picking: Over-pick tol, aug, & annot key.
Community-driven standards to make bioimage data AI-ready & reusable.
👉 www.nature.com/articles/s41... #AI #Bioimaging #FAIRdata

Community-driven standards to make bioimage data AI-ready & reusable.
👉 www.nature.com/articles/s41... #AI #Bioimaging #FAIRdata
We built a phantom cryo-ET dataset (~500 tomograms) + hosted a Kaggle challenge.
The result: community models beat expert tools.
Read more in the @natmethods.nature.com article that just came out:
🔗 doi.org/10.1038/s415...

We built a phantom cryo-ET dataset (~500 tomograms) + hosted a Kaggle challenge.
The result: community models beat expert tools.
Read more in the @natmethods.nature.com article that just came out:
🔗 doi.org/10.1038/s415...
Learn more here: buff.ly/8i8i87U
#SBGrid #SBGridSoftware #StructualBiology

Learn more here: buff.ly/8i8i87U
#SBGrid #SBGridSoftware #StructualBiology
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
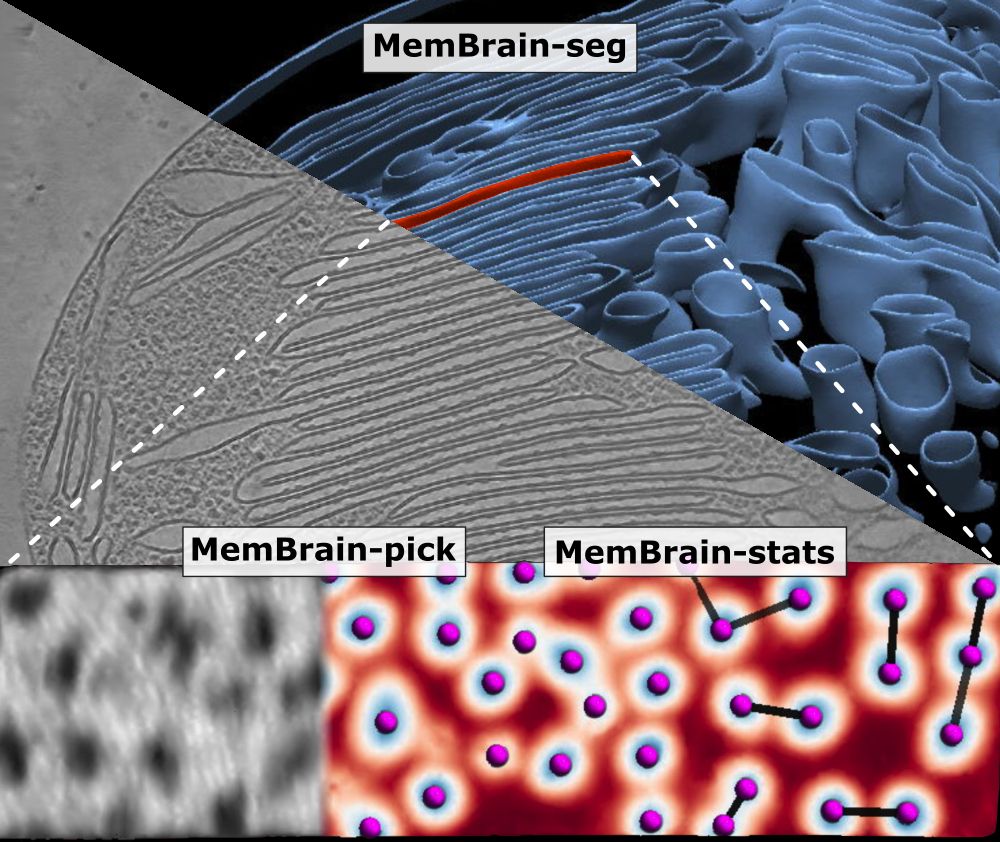
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...
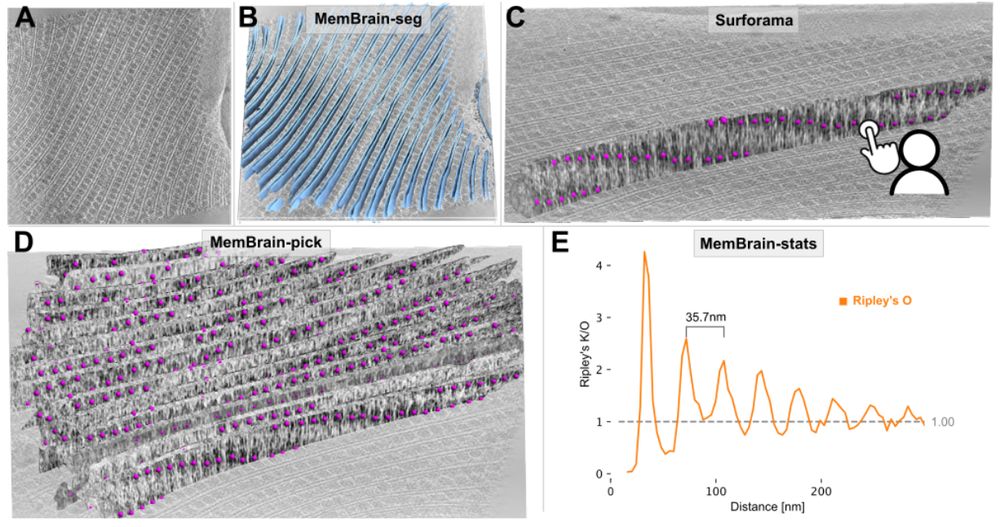
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
kaggle.com/competitions/byu-locating-bacterial-flagellar-motors-2025
kaggle.com/competitions/byu-locating-bacterial-flagellar-motors-2025
Participation exceeded our expectations with 931 Teams from 76 countries competing, generating over 27,000 submissions. For an overview check out our post-competition page: cryoetdataportal.czscience.com/competition
Participation exceeded our expectations with 931 Teams from 76 countries competing, generating over 27,000 submissions. For an overview check out our post-competition page: cryoetdataportal.czscience.com/competition
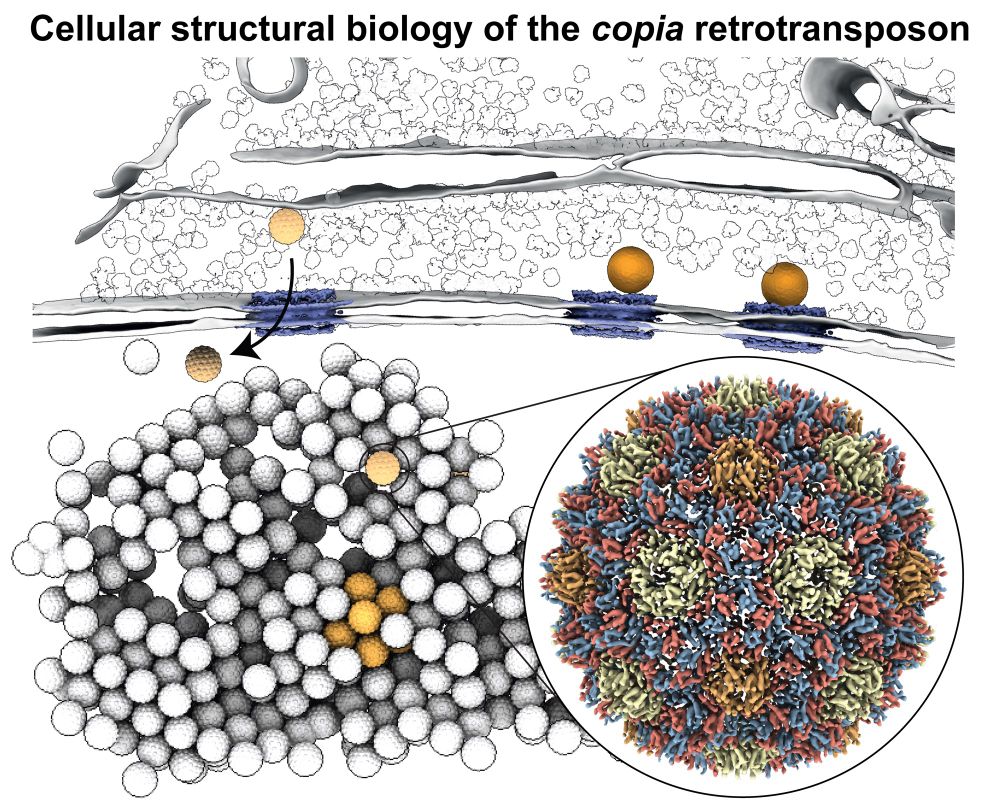
www.ebi.ac.uk/empiar/EMPIA...
A little thread about what's new... 🧵 1/n
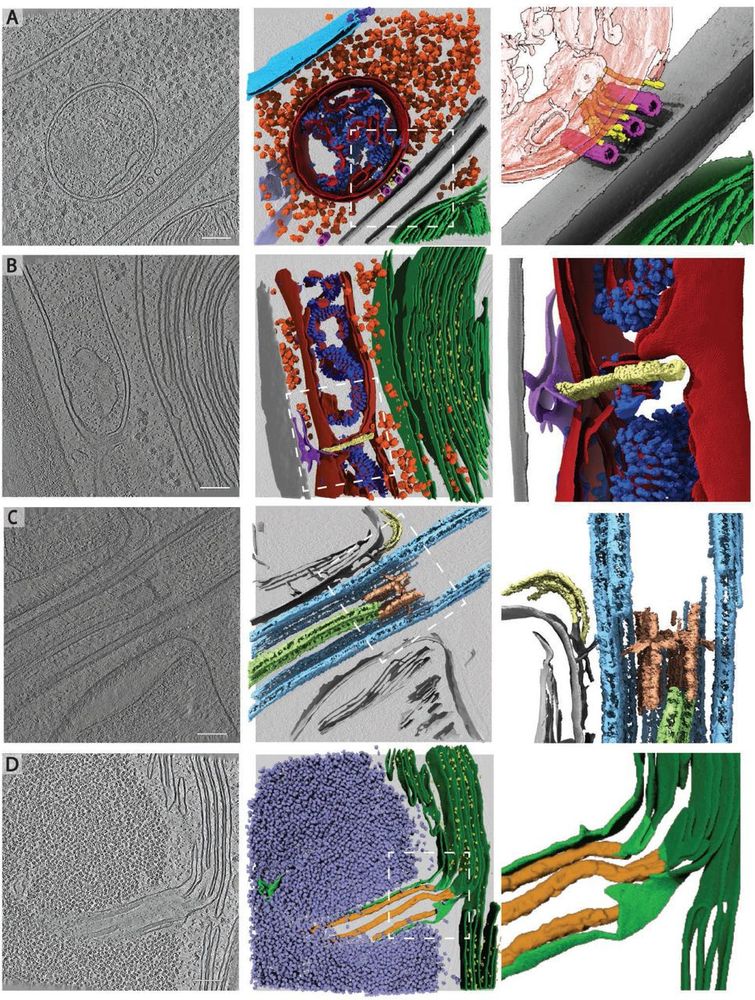
www.ebi.ac.uk/empiar/EMPIA...
A little thread about what's new... 🧵 1/n
Researchers retrained existing AI-based software on over 17,000 microscopy images with over 2 million structures to develop this new model - Segment Anything for Microscopy: www.uni-goettingen.de/en/3240.html...
#NatureMethods research: doi.org/10.1038/s415...


cryoetdataportal.czscience.com/depositions/...

cryoetdataportal.czscience.com/depositions/...

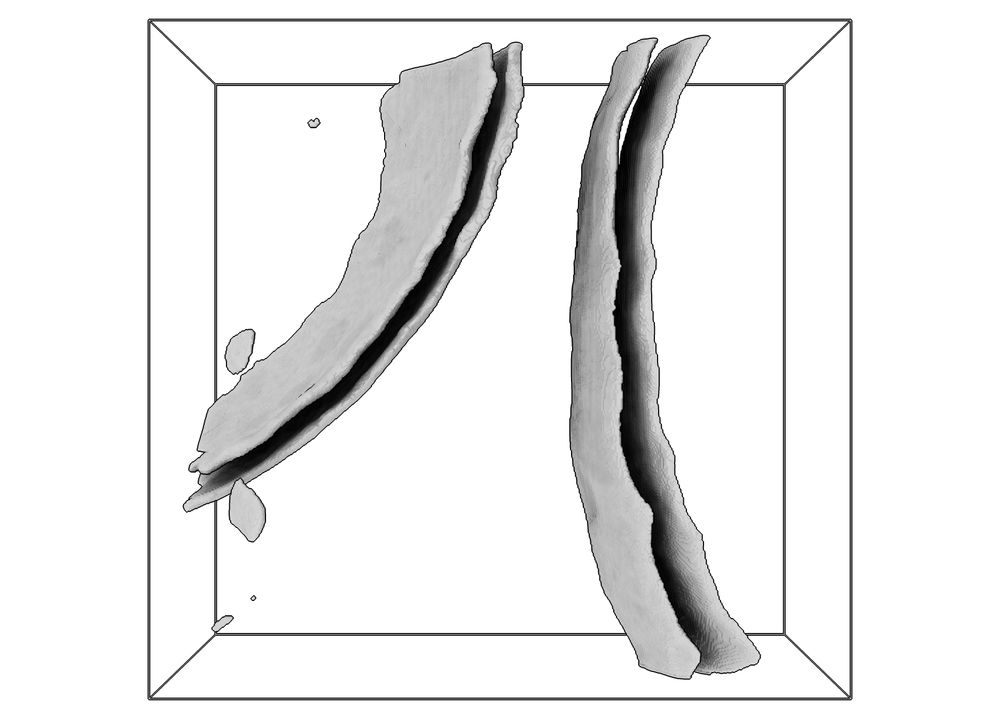
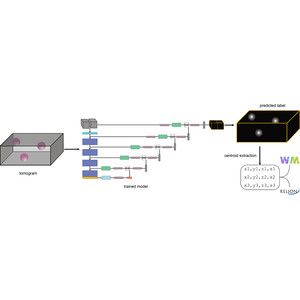
forum.image.sc/t/ngff-0-5-r...
#NGFF #BioImaging #Formats
@openmicroscopy.org @zarr.dev
forum.image.sc/t/ngff-0-5-r...
#NGFF #BioImaging #Formats
@openmicroscopy.org @zarr.dev