Simon Mathis
@simonmathis.bsky.social
PhD student at Uni of Cambridge, UK 🔬 | AI for protein design & engineering 🧬 | biotech & environmental applications 🌱 | enzymes 🏗️
🇦🇹🇨🇭
🇦🇹🇨🇭
Pinned
Simon Mathis
@simonmathis.bsky.social
· Nov 20
🧪 For people interested in AI & enzymes (enzyme engineering, design, discovery, ...), I'm assembling a starter pack for us.
DM if you'd like to be included!
go.bsky.app/MhfaQBh
DM if you'd like to be included!
go.bsky.app/MhfaQBh
Reposted by Simon Mathis
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Accelerating Biomolecular Modeling with AtomWorks and RF3
Deep learning methods trained on protein structure databases have revolutionized biomolecular structure prediction, but developing and training new models remains a considerable challenge. To facilita...
www.biorxiv.org
August 15, 2025 at 5:17 PM
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Reposted by Simon Mathis
RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...

August 15, 2025 at 1:26 PM
RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
Reposted by Simon Mathis
This paper represents a great effort by @roman-bushuiev.bsky.social and his brother @anton-bushuiev.bsky.social. The DreaMS foundation model for mass spectra of small molecules now opens lots of avenues for possible downstream applications. It might be a game changer for computational metabolomics.
Self-supervised learning of molecular representations from millions of tandem mass spectra using DreaMS - @pluskal-lab.org @iocbprague.bsky.social go.nature.com/4k1n5iC

Self-supervised learning of molecular representations from millions of tandem mass spectra using DreaMS - Nature Biotechnology
A transformer model is used to construct the DreaMS Atlas—a molecular network of 201 million MS/MS spectra.
go.nature.com
May 24, 2025 at 8:21 AM
This paper represents a great effort by @roman-bushuiev.bsky.social and his brother @anton-bushuiev.bsky.social. The DreaMS foundation model for mass spectra of small molecules now opens lots of avenues for possible downstream applications. It might be a game changer for computational metabolomics.
Very nice, thoughtful post - I really enjoyed the read @pascalnotin.bsky.social
Even simple methods leveraging these 2 modalities significantly outperform billion-parameter sequence-only models. So, what's next? Better retrieval, advanced multimodal approaches, & alignment. Read more: pascalnotin.substack.com/p/have-we-hi... #BioTech #AI #pLMs
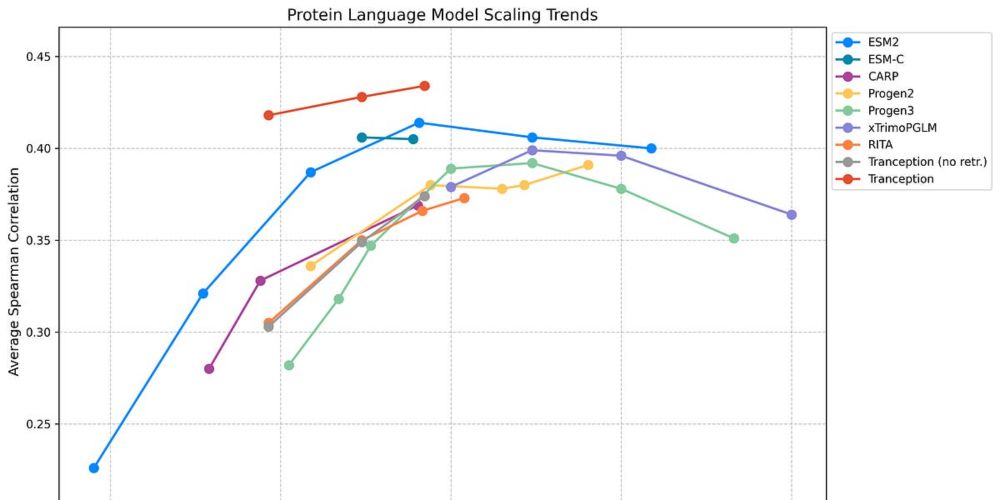
Have We Hit the Scaling Wall for Protein Language Models?
Beyond Scaling: What Truly Works in Protein Fitness Prediction
pascalnotin.substack.com
May 10, 2025 at 10:16 PM
Very nice, thoughtful post - I really enjoyed the read @pascalnotin.bsky.social
Reposted by Simon Mathis
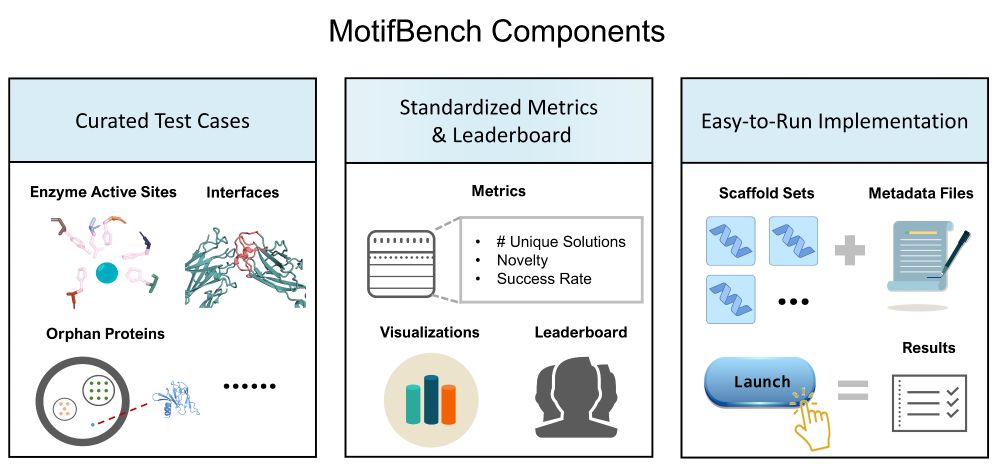
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
February 19, 2025 at 8:50 PM
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
Reposted by Simon Mathis
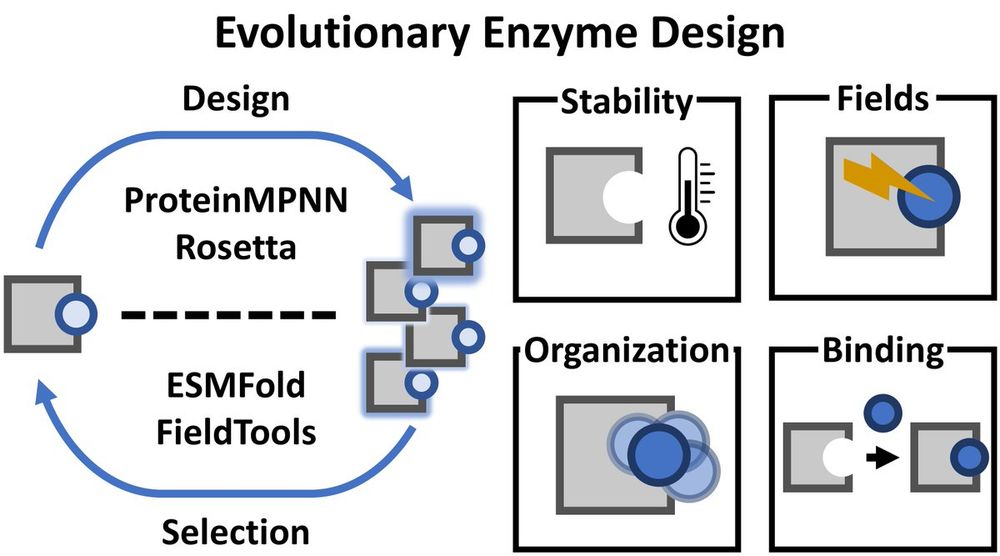
🚨Preprint Alert!🚨
#ProteinDesign is advancing rapidly—wouldn't it be great to seamlessly combine design tools to achieve more than what each can do alone?🤔
Here, we introduce AI.zymes: A modular platform for evolutionary #EnzymeDesign.♻️🖥️
biorxiv.org/content/10.1...
1/🧵
#ProteinDesign is advancing rapidly—wouldn't it be great to seamlessly combine design tools to achieve more than what each can do alone?🤔
Here, we introduce AI.zymes: A modular platform for evolutionary #EnzymeDesign.♻️🖥️
biorxiv.org/content/10.1...
1/🧵
February 12, 2025 at 7:01 PM
🚨Preprint Alert!🚨
#ProteinDesign is advancing rapidly—wouldn't it be great to seamlessly combine design tools to achieve more than what each can do alone?🤔
Here, we introduce AI.zymes: A modular platform for evolutionary #EnzymeDesign.♻️🖥️
biorxiv.org/content/10.1...
1/🧵
#ProteinDesign is advancing rapidly—wouldn't it be great to seamlessly combine design tools to achieve more than what each can do alone?🤔
Here, we introduce AI.zymes: A modular platform for evolutionary #EnzymeDesign.♻️🖥️
biorxiv.org/content/10.1...
1/🧵
Reposted by Simon Mathis
You may have seen a recent pre-print [1] from Jain et al. with strongly worded claims against the experimental results in our DiffDock paper [2]. We initially declined to respond as we saw that this preprint contained falsehoods, misleading comparisons, seemingly deliberate omissions, ...1/n
December 8, 2024 at 9:37 PM
You may have seen a recent pre-print [1] from Jain et al. with strongly worded claims against the experimental results in our DiffDock paper [2]. We initially declined to respond as we saw that this preprint contained falsehoods, misleading comparisons, seemingly deliberate omissions, ...1/n
Reposted by Simon Mathis
Can AI improve the current state of molecular simulation?
www.owlposting.com/p/can-ai-imp...
in my first podcast, I spend 2 hours interviewing Corin Wagen and Ari Wagen, two brothers who are building the next generation of molecular simulation for drug discovery and material science
www.owlposting.com/p/can-ai-imp...
in my first podcast, I spend 2 hours interviewing Corin Wagen and Ari Wagen, two brothers who are building the next generation of molecular simulation for drug discovery and material science

Can AI improve the current state of molecular simulation? (Corin & Ari Wagen, Ep #1)
2.1 hours listening time
www.owlposting.com
December 4, 2024 at 1:50 AM
Can AI improve the current state of molecular simulation?
www.owlposting.com/p/can-ai-imp...
in my first podcast, I spend 2 hours interviewing Corin Wagen and Ari Wagen, two brothers who are building the next generation of molecular simulation for drug discovery and material science
www.owlposting.com/p/can-ai-imp...
in my first podcast, I spend 2 hours interviewing Corin Wagen and Ari Wagen, two brothers who are building the next generation of molecular simulation for drug discovery and material science
Reposted by Simon Mathis
We are hiring (resharing appreciated)!
Given recent successful grant applications (I got my SNSF Starting Grant 🚀), we are extending the LIAC team with multiple openings (PhD/postdoc) for 2025.
Apply now (deadline: December 20th) by filling in this form: forms.fillout.com/t/eq5ADAw3kkus.
#ChemSky
Given recent successful grant applications (I got my SNSF Starting Grant 🚀), we are extending the LIAC team with multiple openings (PhD/postdoc) for 2025.
Apply now (deadline: December 20th) by filling in this form: forms.fillout.com/t/eq5ADAw3kkus.
#ChemSky



December 2, 2024 at 10:33 AM
We are hiring (resharing appreciated)!
Given recent successful grant applications (I got my SNSF Starting Grant 🚀), we are extending the LIAC team with multiple openings (PhD/postdoc) for 2025.
Apply now (deadline: December 20th) by filling in this form: forms.fillout.com/t/eq5ADAw3kkus.
#ChemSky
Given recent successful grant applications (I got my SNSF Starting Grant 🚀), we are extending the LIAC team with multiple openings (PhD/postdoc) for 2025.
Apply now (deadline: December 20th) by filling in this form: forms.fillout.com/t/eq5ADAw3kkus.
#ChemSky
For some more guidance on how to use this, Martin Buttenschön wrote a nice blogpost: www.blopig.com/blog/2024/11...
November 29, 2024 at 10:27 AM
For some more guidance on how to use this, Martin Buttenschön wrote a nice blogpost: www.blopig.com/blog/2024/11...
Reposted by Simon Mathis
Check out our MassSpecGym dataset on @polarishq.bsky.social. 🤩
Wow! 🤩
This may be the most carefully documented dataset on @polarishq.bsky.social. Great work by @roman-bushuiev.bsky.social!
Do I have any #MassSpec researchers in my 🦋 network yet? I would love to hear what you think! github.com/polaris-hub/...
#mass #spectrometry #dataset #benchmark
This may be the most carefully documented dataset on @polarishq.bsky.social. Great work by @roman-bushuiev.bsky.social!
Do I have any #MassSpec researchers in my 🦋 network yet? I would love to hear what you think! github.com/polaris-hub/...
#mass #spectrometry #dataset #benchmark

massspecgym
roman-bushuiev
polarishub.io
November 27, 2024 at 8:18 AM
Check out our MassSpecGym dataset on @polarishq.bsky.social. 🤩
Reposted by Simon Mathis
Conformational dynamics smoothen the fitness landscapes of enzymes. Who knew? 🧪🧶

November 27, 2024 at 4:17 AM
Conformational dynamics smoothen the fitness landscapes of enzymes. Who knew? 🧪🧶
Reposted by Simon Mathis
✨ wheeeeee we raised a series B
AI-powered automated protein optimization: matches or exceeds human performance, works without human intervention. It's pretty cool :D
(PS if you're seriously good at ML eng then we're hiring)
www.cradle.bio/blog/series-b
AI-powered automated protein optimization: matches or exceeds human performance, works without human intervention. It's pretty cool :D
(PS if you're seriously good at ML eng then we're hiring)
www.cradle.bio/blog/series-b

Cradle – Cradle raises $73M Series B to Put AI-Powered Protein Engineering in Every Lab
News, updates, tutorials, and more from the makers of Cradle
www.cradle.bio
November 26, 2024 at 8:50 AM
✨ wheeeeee we raised a series B
AI-powered automated protein optimization: matches or exceeds human performance, works without human intervention. It's pretty cool :D
(PS if you're seriously good at ML eng then we're hiring)
www.cradle.bio/blog/series-b
AI-powered automated protein optimization: matches or exceeds human performance, works without human intervention. It's pretty cool :D
(PS if you're seriously good at ML eng then we're hiring)
www.cradle.bio/blog/series-b
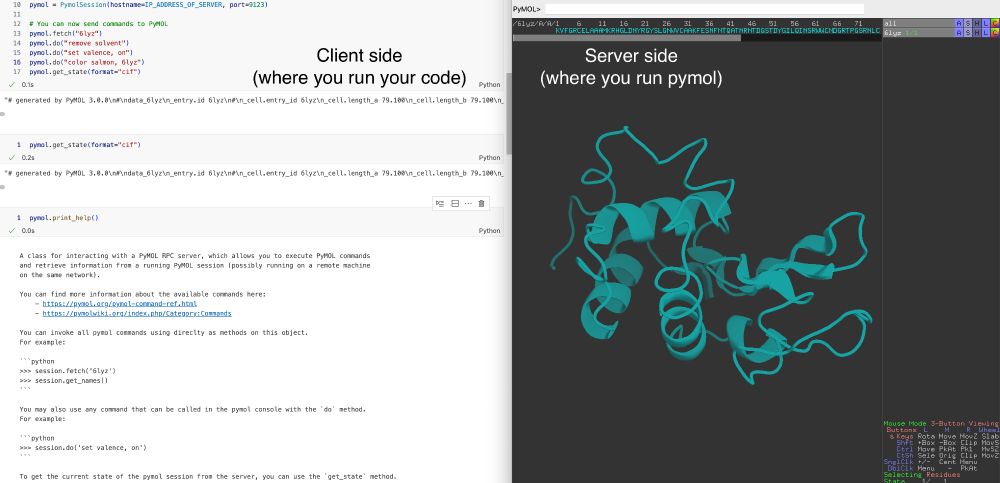
A weekend project from a while back -- this little package (with no dependencies) allows you to interact with pymol remotely.
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
November 25, 2024 at 2:50 PM
A weekend project from a while back -- this little package (with no dependencies) allows you to interact with pymol remotely.
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
Nice overview, thank you @harrisbio.bsky.social !
Now I can share external links without the posts being down regulated - I thought it would share this again!:)
I have compiled a list of now 100+ companies in the 'TechBio' space into a fully open database for the community.
Find here and please share if you like
open.substack.com/pub/harrisbi...
I have compiled a list of now 100+ companies in the 'TechBio' space into a fully open database for the community.
Find here and please share if you like
open.substack.com/pub/harrisbi...

November 21, 2024 at 12:25 PM
Nice overview, thank you @harrisbio.bsky.social !
🧪 For people interested in AI & enzymes (enzyme engineering, design, discovery, ...), I'm assembling a starter pack for us.
DM if you'd like to be included!
go.bsky.app/MhfaQBh
DM if you'd like to be included!
go.bsky.app/MhfaQBh
November 20, 2024 at 10:29 AM
🧪 For people interested in AI & enzymes (enzyme engineering, design, discovery, ...), I'm assembling a starter pack for us.
DM if you'd like to be included!
go.bsky.app/MhfaQBh
DM if you'd like to be included!
go.bsky.app/MhfaQBh
Thank you for organising this @kevinkaichuang.bsky.social
Two BioML starter packs now:
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
November 20, 2024 at 9:37 AM
Thank you for organising this @kevinkaichuang.bsky.social

