Pascal Notin
@pascalnotin.bsky.social
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique
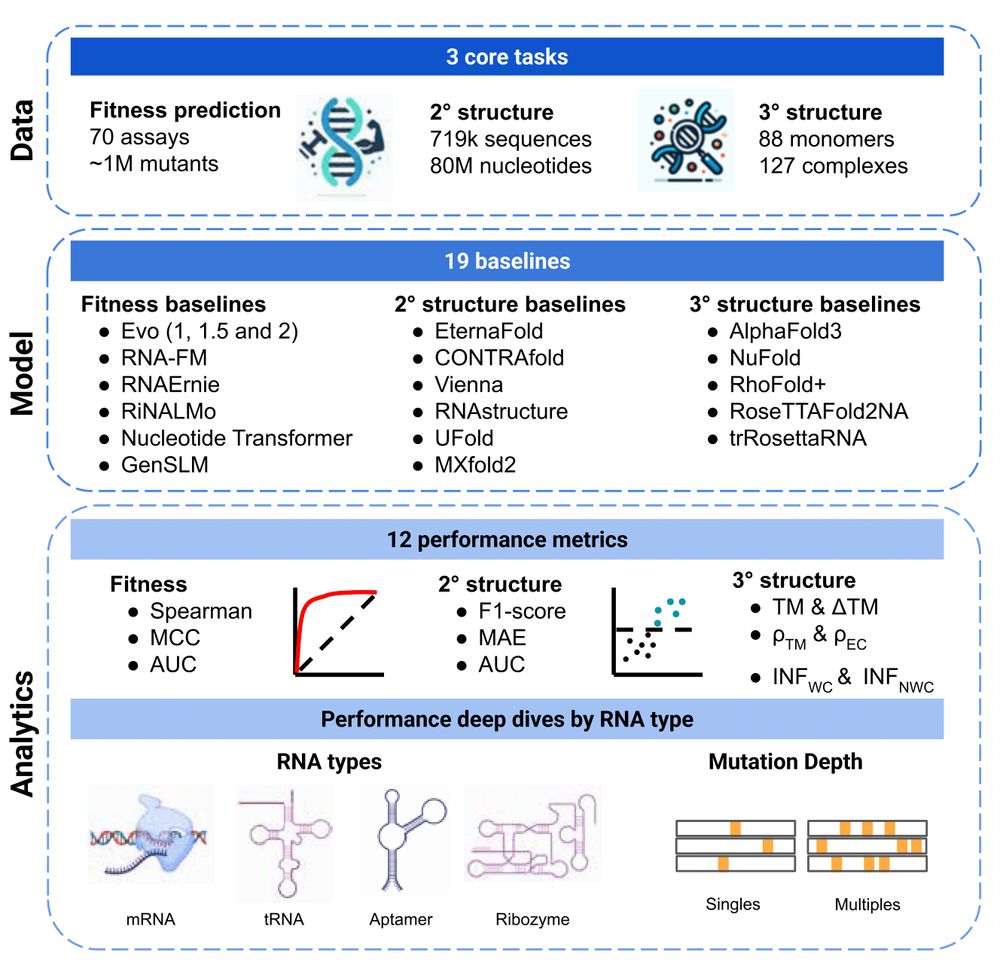
🚨 New paper 🚨 RNA modeling just got its own Gym! 🏋️ Introducing RNAGym, large-scale benchmarks for RNA fitness and structure prediction.
🧵 1/9
🧵 1/9
June 18, 2025 at 7:35 PM
🚨 New paper 🚨 RNA modeling just got its own Gym! 🏋️ Introducing RNAGym, large-scale benchmarks for RNA fitness and structure prediction.
🧵 1/9
🧵 1/9
Reposted by Pascal Notin
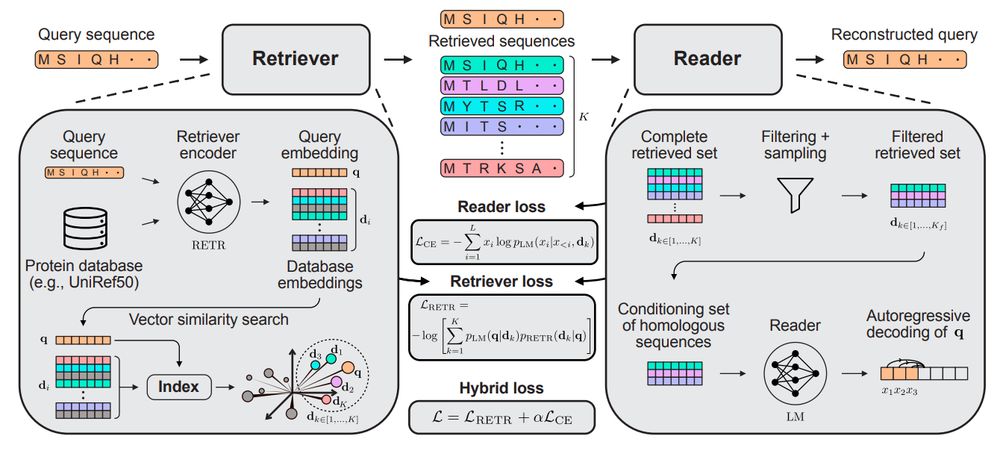
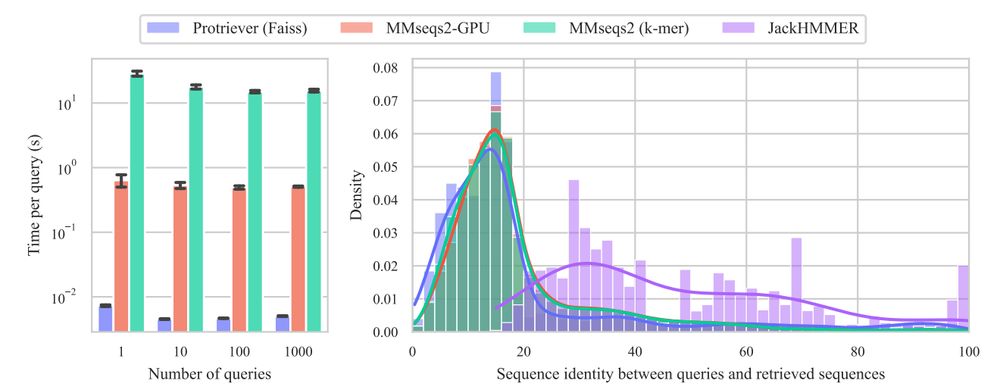
End-to-end differentiable homology search for protein fitness prediction.
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
June 11, 2025 at 7:00 PM
End-to-end differentiable homology search for protein fitness prediction.
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
Reposted by Pascal Notin
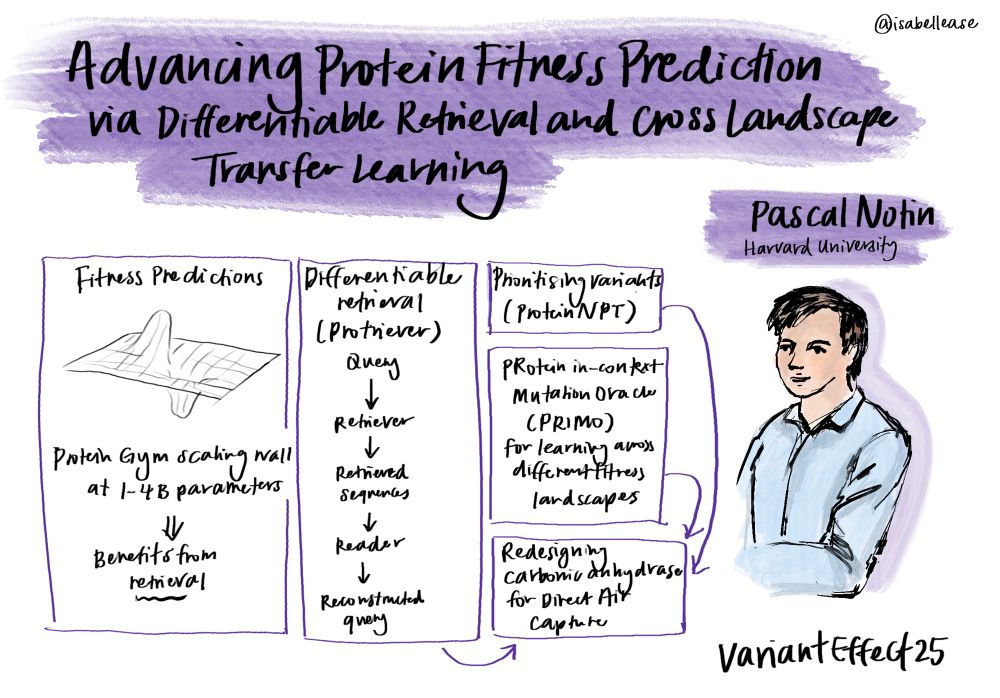
Have we hit a "scaling wall" for protein language models? 🤔 Our latest ProteinGym v1.3 release suggests that for zero-shot fitness prediction, simply making pLMs bigger isn't better beyond 1-4B parameters. The winning strategy? Combining MSAs & structure in multimodal models!
May 8, 2025 at 12:29 AM
Have we hit a "scaling wall" for protein language models? 🤔 Our latest ProteinGym v1.3 release suggests that for zero-shot fitness prediction, simply making pLMs bigger isn't better beyond 1-4B parameters. The winning strategy? Combining MSAs & structure in multimodal models!
Reposted by Pascal Notin
Large-scale discovery, analysis, and design of protein energy landscapes https://www.biorxiv.org/content/10.1101/2025.03.20.644235v1
March 25, 2025 at 2:47 PM
Large-scale discovery, analysis, and design of protein energy landscapes https://www.biorxiv.org/content/10.1101/2025.03.20.644235v1