I’m a former NIH PI. Happy to refer you to a law firm that has successfully sued the govt many times!
Pls repost 🙏❤️
I’m a former NIH PI. Happy to refer you to a law firm that has successfully sued the govt many times!
Pls repost 🙏❤️
grants.nih.gov/grants/guide...

grants.nih.gov/grants/guide...
www.nature.com/articles/s41...
Unfortunately, it makes several misleading claims that we disagree with. We will address these technical & other issues with this work over the next few months.

www.nature.com/articles/s41...
Unfortunately, it makes several misleading claims that we disagree with. We will address these technical & other issues with this work over the next few months.
arxiv.org/abs/2411.11158

arxiv.org/abs/2411.11158
Paper: www.biorxiv.org/content/10.1...

Paper: www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
1/7
openreview.net/forum?id=H7m...

1/7
openreview.net/forum?id=H7m...
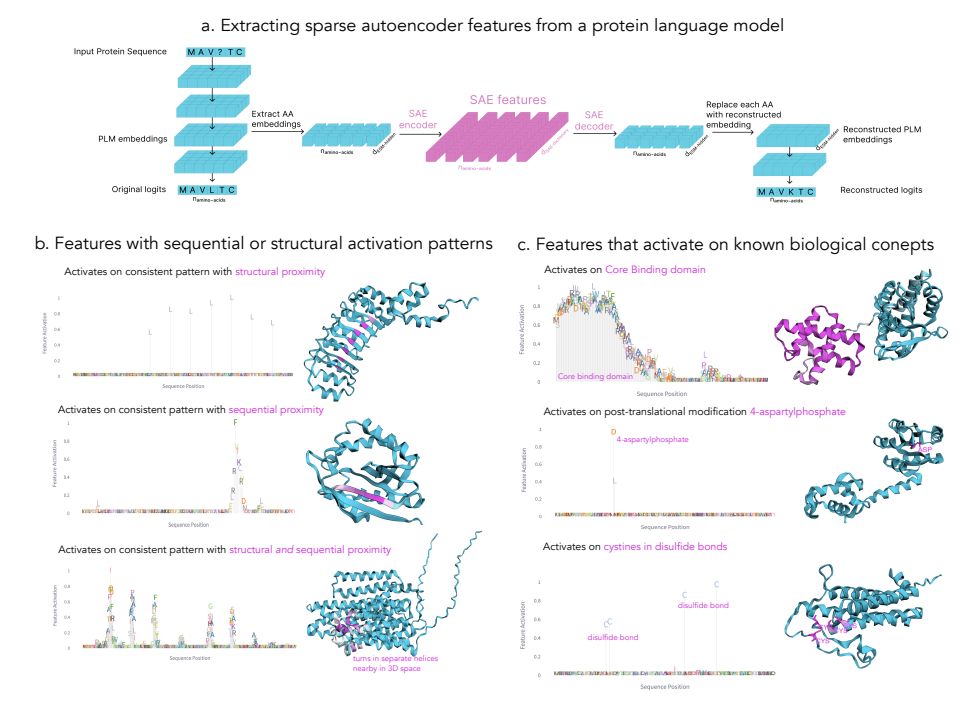
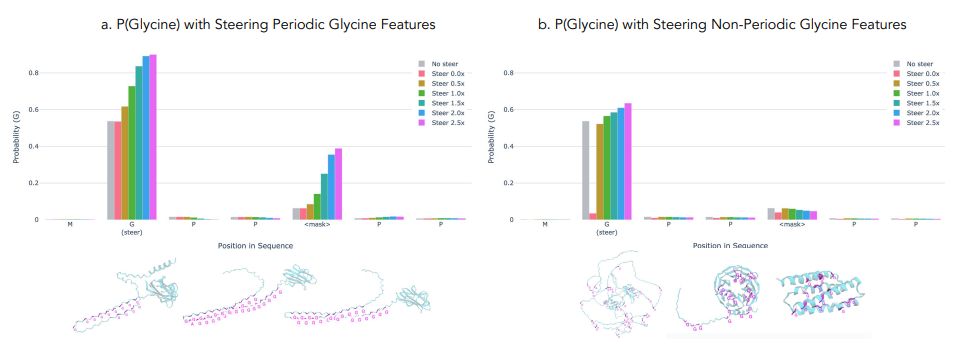
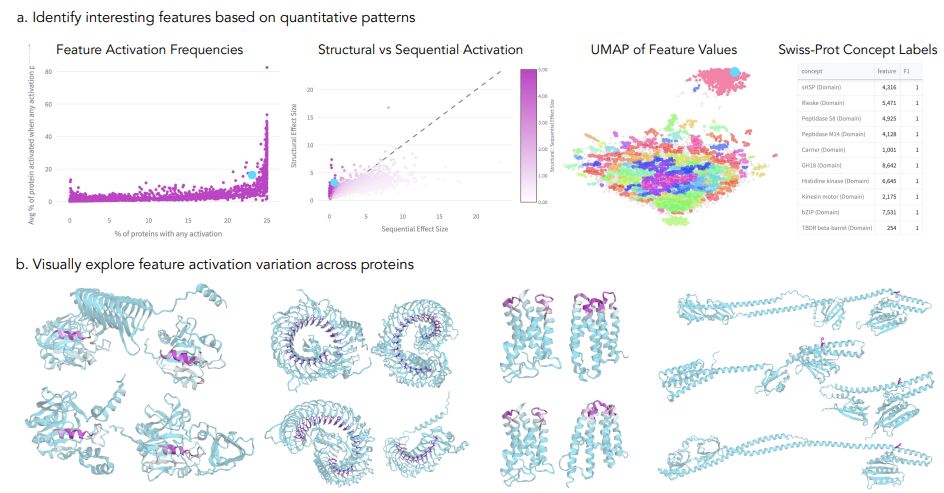
Blog-post jclinic.mit.edu/boltz-1/
Technical report: t.co/px0spnTA0S
Model and code: github.com/jwohlwend/boltz
Blog-post jclinic.mit.edu/boltz-1/
Technical report: t.co/px0spnTA0S
Model and code: github.com/jwohlwend/boltz
TCR-TRANSLATE: Conditional Generation of Real Antigen Specific T-cell Receptor Sequences
www.biorxiv.org/content/10.1...
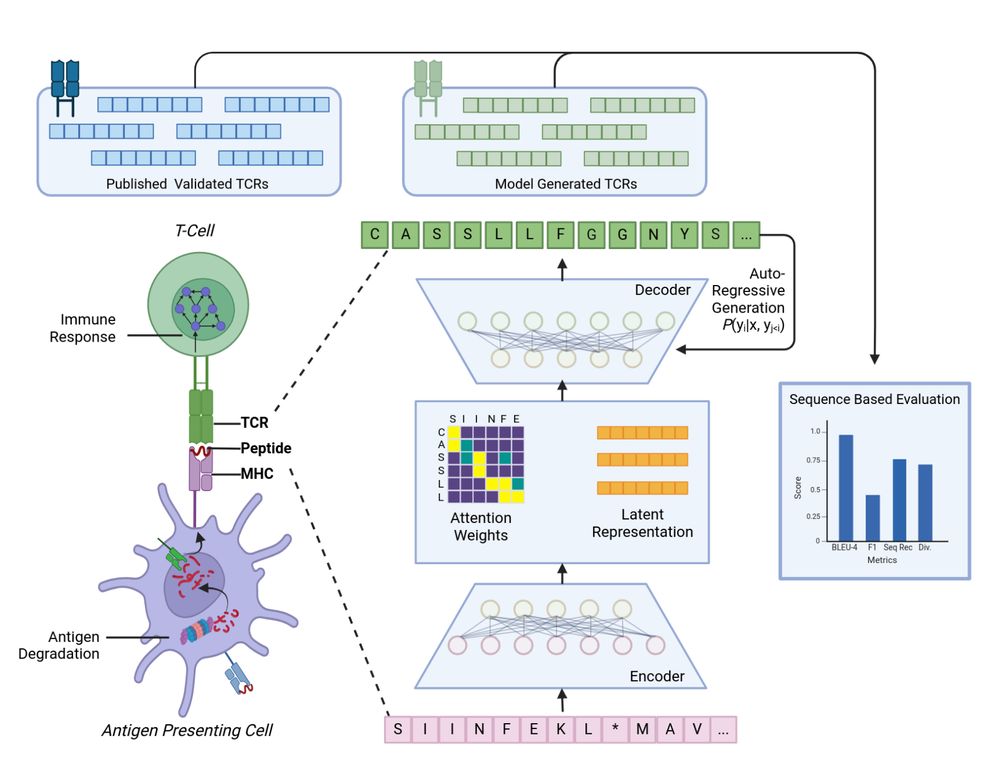
TCR-TRANSLATE: Conditional Generation of Real Antigen Specific T-cell Receptor Sequences
www.biorxiv.org/content/10.1...


