
Eric Kernfeld
@ekernf01.bsky.social
Statistician and computational biologist; uw alum; jhu student. He/him. http://ekernf01.github.io
This is so friggin cool. You don't have to rewrite all that FORTRAN to do autodiff on it. "Enzyme"
Found the paper
arxiv.org/abs/2010.01709
arxiv.org/abs/2010.01709

Instead of Rewriting Foreign Code for Machine Learning, Automatically Synthesize Fast Gradients
Applying differentiable programming techniques and machine learning algorithms to foreign programs requires developers to either rewrite their code in a machine learning framework, or otherwise provid...
arxiv.org
October 12, 2025 at 9:00 PM
This is so friggin cool. You don't have to rewrite all that FORTRAN to do autodiff on it. "Enzyme"
I love using the UKB RAP. As soon as I see that £, my brain goes "I shall keep-safe this file upon the clouds and it shall cost you nigh thruppence fortnightly." innit
September 19, 2025 at 10:49 PM
I love using the UKB RAP. As soon as I see that £, my brain goes "I shall keep-safe this file upon the clouds and it shall cost you nigh thruppence fortnightly." innit
For people that have tried to hire someone recently, did you encounter this problem?
www.linkedin.com/posts/olga-v...
www.linkedin.com/posts/olga-v...

Failed to weed out LLM-generated applications for bioinformatics role | Olga Sazonova, Ph.D. posted on the topic | LinkedIn
Well, that's it. Hiring is f*cked. I'm just opened a contract role for a bioinformatics project. I know it's a wild market at the moment - each open role gets flooded with resumes, making it hard to separate qualified candidates from the noise. I'm convinced LLMs are partially to blame. So I devised a process to select qualified, committed candidates and avoid generic applications. Instead of resumes and cover letters, I created an intake questionnaire requiring bespoke effort to reduce cookie-cutter applications. The questions focused on specific scenarios from past experience, and therefore shouldn't be directly outsourced to chatbots. I also accidentally made the application link hard to access, but decided not to fix it. After all, computational biology often requires hacking and clever work-arounds. Finally, I included an honor code asking people not to use LLMs. Initially, I felt good about my approach. I had 20 applications after two days, and the candidates were differentiating themselves: A few didn't answer all questions, a few didn't have the right background, and several candidates provided well-written, topical, plausible answers. It was only after I read a few of these "high signal" applications in a row that the alarm bells started ringing: - Multiple candidates highlighted the same methodological paper - Many cited required skills in the exact same order - A high percentage reported identical troubleshooting scenarios involving differential gene expression studies with lab-derived batch effects The nail in the coffin was repeated phrases like "my initial hypothesis was a bug in [popular tool]" and "the spurious result disappeared." Clearly, candidates were using LLMs. Instead of learning about fitness for the role, I was learning how LLMs answer my "clever" questions. So...I failed. My approach was naive, perhaps even hypocritical. After all, I use LLMs for technical documentation myself. Still, it's really disappointing. I'm no closer to a hiring process that identifies qualified, motivated, and trustworthy candidates for remote work. Now what, y'all? | 109 comments on LinkedIn
www.linkedin.com
August 30, 2025 at 1:54 PM
For people that have tried to hire someone recently, did you encounter this problem?
www.linkedin.com/posts/olga-v...
www.linkedin.com/posts/olga-v...
All parrots are stochastic.
August 16, 2025 at 12:21 PM
All parrots are stochastic.
Job Search Recap
ekernf01.github.io/job_search_2...
ekernf01.github.io/job_search_2...
![Sankey diagram of Eric's job search efforts and opportunities. Source data:
Months unemployed [1] Biking
Months unemployed [1] Stressing
Months unemployed [1] Applying
Months unemployed [2] Networking
Months unemployed [0] Waiting to joke about "came in a fluffer"
Jobs applied to (no referral) [64] No follow up
Jobs applied to (with referral) [4] No follow up
Jobs applied to (with referral) [3] Interview
Jobs applied to (no referral) [1] Interview
Info interviews conducted [30] No follow up
Info interviews conducted [2] Interview
Credible unsolicited inquiries [2] Interview
Credible unsolicited inquiries [3] Location mismatch
Interview [2] Job offer](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:5ftexbig72jeep64y6wzkj4z/bafkreibw2gfme7kbz4zyj47nh44b52zirtrprbtpti24svg534vwt44pr4@jpeg)
August 13, 2025 at 3:26 PM
Job Search Recap
ekernf01.github.io/job_search_2...
ekernf01.github.io/job_search_2...
I am proud to announce a lot of personal progress from this spring and summer. 🧵
August 13, 2025 at 3:20 PM
I am proud to announce a lot of personal progress from this spring and summer. 🧵
In October 2024, I twote that "something is deeply wrong" with what we now call virtual cell models. A lot has happened since then. How am I updating? New blog post: ekernf01.github.io/virtual-cell...
A recap of virtual cell releases circa June 2025
In October 2024, I twote that “something is deeply wrong” with what we now call virtual cell models. A lot has happened since then: modelers are advancing new architectures and mining new sources of i...
ekernf01.github.io
July 27, 2025 at 11:48 PM
In October 2024, I twote that "something is deeply wrong" with what we now call virtual cell models. A lot has happened since then. How am I updating? New blog post: ekernf01.github.io/virtual-cell...
Reposted by Eric Kernfeld
The biggest challenge for AI in biology isn't just models, it's the data used to train them. Standard biological data isn't built for AI. To unlock generative AI for drug discovery, we must rethink how we generate and capture data. 1/

July 22, 2025 at 12:30 PM
The biggest challenge for AI in biology isn't just models, it's the data used to train them. Standard biological data isn't built for AI. To unlock generative AI for drug discovery, we must rethink how we generate and capture data. 1/
Reposted by Eric Kernfeld
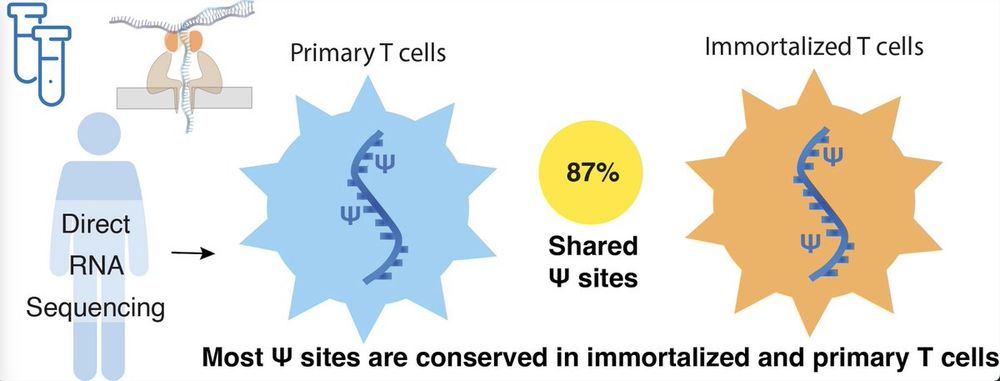
1/🧵
🚨 New paper published in RNA!
Scientists often say anecdotally that RNA modifications are disrupted in immortalized cells — but no one’s really tested it.
So we did: ψ-mapping in primary T cells vs. Jurkat cells using direct RNA-seq.
📄 tinyurl.com/TCellPsiRNA
#nanopore #RNA #pseudouridine
🚨 New paper published in RNA!
Scientists often say anecdotally that RNA modifications are disrupted in immortalized cells — but no one’s really tested it.
So we did: ψ-mapping in primary T cells vs. Jurkat cells using direct RNA-seq.
📄 tinyurl.com/TCellPsiRNA
#nanopore #RNA #pseudouridine
July 17, 2025 at 5:22 PM
1/🧵
🚨 New paper published in RNA!
Scientists often say anecdotally that RNA modifications are disrupted in immortalized cells — but no one’s really tested it.
So we did: ψ-mapping in primary T cells vs. Jurkat cells using direct RNA-seq.
📄 tinyurl.com/TCellPsiRNA
#nanopore #RNA #pseudouridine
🚨 New paper published in RNA!
Scientists often say anecdotally that RNA modifications are disrupted in immortalized cells — but no one’s really tested it.
So we did: ψ-mapping in primary T cells vs. Jurkat cells using direct RNA-seq.
📄 tinyurl.com/TCellPsiRNA
#nanopore #RNA #pseudouridine
BLOG ALERT! 🧬 If you do RNA-seq, ATAC-seq, ChIP-seq, or modeling thereof, you may be overlooking LD score regression methods. These should be standard tools to study genes, variants, and regions en masse, but they are hard to understand. I wrote intros:

June 26, 2025 at 1:03 PM
BLOG ALERT! 🧬 If you do RNA-seq, ATAC-seq, ChIP-seq, or modeling thereof, you may be overlooking LD score regression methods. These should be standard tools to study genes, variants, and regions en masse, but they are hard to understand. I wrote intros:
Fast-followers are more lucrative than first-in-class drugs. centuryofbio.com/p/commoditiz...
How, then, can companies protect investments in target discovery?
Code obfuscators protect source code from reverse engineering.
Could mechanism obfuscators protect drug target identities?
How, then, can companies protect investments in target discovery?
Code obfuscators protect source code from reverse engineering.
Could mechanism obfuscators protect drug target identities?

On Modality Commoditization
And what might come next
centuryofbio.com
June 25, 2025 at 1:38 PM
Fast-followers are more lucrative than first-in-class drugs. centuryofbio.com/p/commoditiz...
How, then, can companies protect investments in target discovery?
Code obfuscators protect source code from reverse engineering.
Could mechanism obfuscators protect drug target identities?
How, then, can companies protect investments in target discovery?
Code obfuscators protect source code from reverse engineering.
Could mechanism obfuscators protect drug target identities?
Help me understand the definition of genetic covariance from the xt-ldsc paper. If beta is an argmax, then isn't cbeta also an argmax for any positive scalar c?

June 23, 2025 at 6:04 PM
Help me understand the definition of genetic covariance from the xt-ldsc paper. If beta is an argmax, then isn't cbeta also an argmax for any positive scalar c?
Is anyone making giant post-perturbation cDNA libraries and sticking them in the freezer while waiting for sequencing costs to drop below a certain point? editlife.substack.com/p/part-1-the...

Part 1. The DNA Sequencing Arms Race: The Fight to Dethrone Illumina
Multi-part series exploring the tech and politics around short/long-read sequencing advances
editlife.substack.com
June 22, 2025 at 12:26 PM
Is anyone making giant post-perturbation cDNA libraries and sticking them in the freezer while waiting for sequencing costs to drop below a certain point? editlife.substack.com/p/part-1-the...
Terrific opportunity. If I were not committed to the east coast for personal reasons, I would apply to this immediately.
@saramostafavi.bsky.social (@Genentech) & I (@Stanford) r excited to announce co-advised postdoc positions for candidates with deep expertise in ML for bio (especially sequence to function models, causal perturbational models & single cell models). See details below. Pls RT 1/

June 19, 2025 at 9:10 PM
Terrific opportunity. If I were not committed to the east coast for personal reasons, I would apply to this immediately.
It's really cool to live in a time when a paper on differential expression testing will casually drop a new 160,000-cell perturb-seq experiment.
www.ncbi.nlm.nih.gov/geo/query/ac...
www.ncbi.nlm.nih.gov/geo/query/ac...
GEO Accession viewer
NCBI's Gene Expression Omnibus (GEO) is a public archive and resource for gene expression data.
www.ncbi.nlm.nih.gov
June 19, 2025 at 4:34 PM
It's really cool to live in a time when a paper on differential expression testing will casually drop a new 160,000-cell perturb-seq experiment.
www.ncbi.nlm.nih.gov/geo/query/ac...
www.ncbi.nlm.nih.gov/geo/query/ac...
If you see this, I want your answer to this question from @alz_zyd_ on twitter:
"What is the largest scope/time project, of any kind, you've accomplished in your life so far?"
"What is the largest scope/time project, of any kind, you've accomplished in your life so far?"
June 17, 2025 at 6:06 PM
If you see this, I want your answer to this question from @alz_zyd_ on twitter:
"What is the largest scope/time project, of any kind, you've accomplished in your life so far?"
"What is the largest scope/time project, of any kind, you've accomplished in your life so far?"
Reposted by Eric Kernfeld
EPIGENETIC HULK READY TO SMASH AGAIN! GET IN LOSERS!
June 13, 2025 at 4:30 AM
EPIGENETIC HULK READY TO SMASH AGAIN! GET IN LOSERS!
Reposted by Eric Kernfeld
Thrilled to announce that I am joining DTU in Copenhagen in the fall, as an assistant professor of chemistry.
My research group will focus on fundamental methodology in machine learning for molecules.
My research group will focus on fundamental methodology in machine learning for molecules.
May 29, 2025 at 5:04 PM
Thrilled to announce that I am joining DTU in Copenhagen in the fall, as an assistant professor of chemistry.
My research group will focus on fundamental methodology in machine learning for molecules.
My research group will focus on fundamental methodology in machine learning for molecules.
Another case showing we must be wary of Gell-Mann amnesia in evaluating molecular bio applications of AI.
rachel.fast.ai/posts/2025-0...
rachel.fast.ai/posts/2025-0...

Rachel Thomas, PhD - Deep learning gets the glory, deep fact checking gets ignored
an AI researcher going back to school for immunology
rachel.fast.ai
June 5, 2025 at 9:36 PM
Another case showing we must be wary of Gell-Mann amnesia in evaluating molecular bio applications of AI.
rachel.fast.ai/posts/2025-0...
rachel.fast.ai/posts/2025-0...
Reposted by Eric Kernfeld
1/ DNA sequence models like Borzoi predict gene expression and variant effects across tissues — but how can someone adapt the model to a custom experiment? @drkbio.bsky.social, Johannes Linder and I propose a solution via parameter-efficient fine-tuning (PEFT).
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Parameter-Efficient Fine-Tuning of a Supervised Regulatory Sequence Model
DNA sequence deep learning models accurately predict epigenetic and transcriptional profiles, enabling analysis of gene regulation and genetic variant effects. While large-scale training models like E...
www.biorxiv.org
June 2, 2025 at 8:09 PM
1/ DNA sequence models like Borzoi predict gene expression and variant effects across tissues — but how can someone adapt the model to a custom experiment? @drkbio.bsky.social, Johannes Linder and I propose a solution via parameter-efficient fine-tuning (PEFT).
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Eric Kernfeld
Are you an early-career researcher working on the computational analysis of population-level human genetics data? We want to hear from you about if, how, and why you use population descriptors in your research! Fill out our short survey: forms.gle/SCiNUq71wgi5...

June 2, 2025 at 5:32 PM
Are you an early-career researcher working on the computational analysis of population-level human genetics data? We want to hear from you about if, how, and why you use population descriptors in your research! Fill out our short survey: forms.gle/SCiNUq71wgi5...
Reposted by Eric Kernfeld
The duplication crisis: the other replication crisis - www.worksinprogress.news/p/the-duplic...

The duplication crisis: the other replication crisis
How bad publishing incentives hinder long-term thinking in computational biology research
www.worksinprogress.news
June 2, 2025 at 7:41 PM
The duplication crisis: the other replication crisis - www.worksinprogress.news/p/the-duplic...
new advance in classifying species-of-origin for long reads. Potentially very valuable for diagnostics!! But hot damn, look at that memory consumption.
1/5 We introduce Movi Color, led by Steven Tan (a brilliant undergrad member of Langmead lab) for taxonomic and multi-class classification. It uses a full-text index based on the move structure and does not rely on predefined values (like k-mer length) for index building.
github.com/mohsenzakeri...
github.com/mohsenzakeri...
May 29, 2025 at 2:53 PM
new advance in classifying species-of-origin for long reads. Potentially very valuable for diagnostics!! But hot damn, look at that memory consumption.
Wake up, babe. New cat coat color mechanism just dropped. www.theguardian.com/us-news/2025...

Scientists solve the mystery of ginger cats – helped by hundreds of cat owners
Discovery has implications for ‘all cells and tissues’ and research bridged gap between scientists and non-scientists
www.theguardian.com
May 28, 2025 at 5:47 PM
Wake up, babe. New cat coat color mechanism just dropped. www.theguardian.com/us-news/2025...


