
Check out 'one-pot' PASTA recipe 🔥🍝: www.biorxiv.org/content/10.1101/2025.09.10.675267v1
We combined a unique mix of CRISPR knock-in with serine integrases for easy-to-adopt
𝐏rogrammable
𝐀nd
𝐒ite-specific
𝐓ransgene
𝐀ddition
🧪

Check out 'one-pot' PASTA recipe 🔥🍝: www.biorxiv.org/content/10.1101/2025.09.10.675267v1
We combined a unique mix of CRISPR knock-in with serine integrases for easy-to-adopt
𝐏rogrammable
𝐀nd
𝐒ite-specific
𝐓ransgene
𝐀ddition
🧪

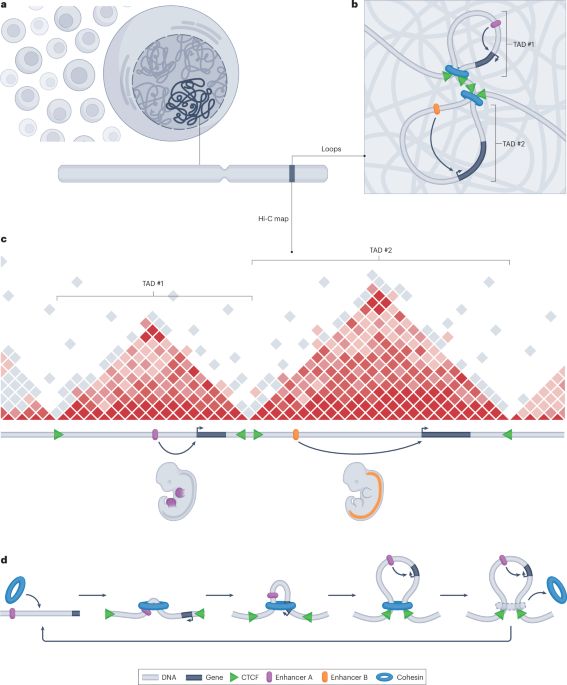
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
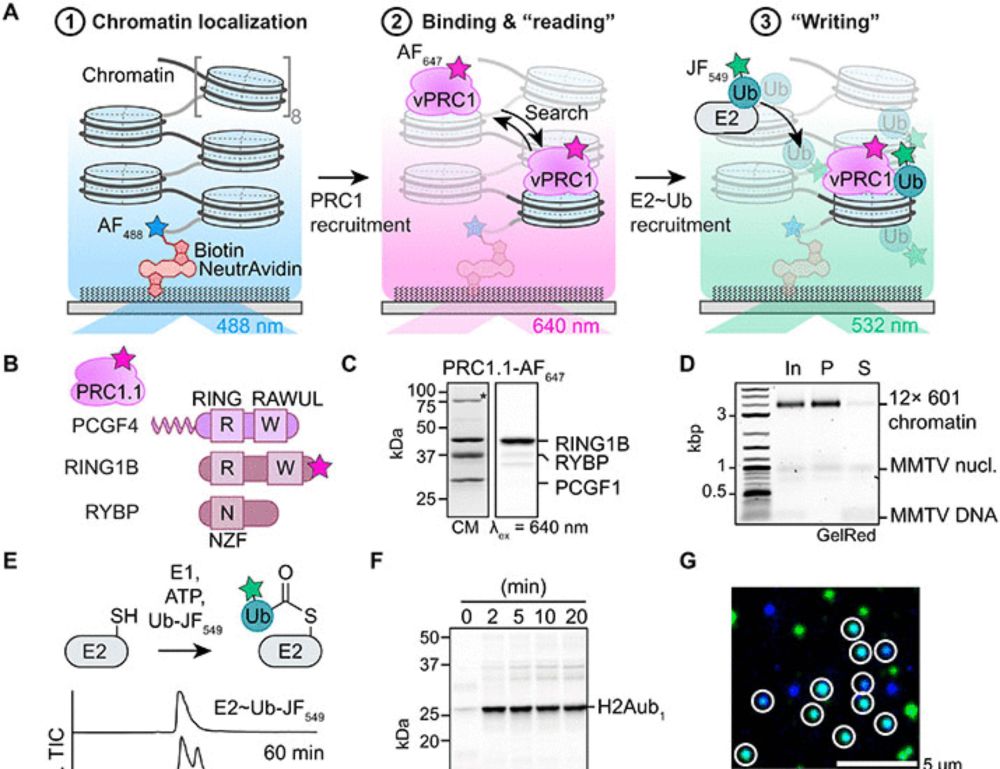
👉 Read the full article here: https://doi.org/10.1093/nar/gkaf056

👉 Read the full article here: https://doi.org/10.1093/nar/gkaf056
academic.oup.com/nar/advance-...

academic.oup.com/nar/advance-...

