Genome Architecture | Gene regulation | Live-cell imaging | Genomics
I am delighted that our paper exploring the impact of Neanderthal-derived variants on the activity of a disease-associated craniofacial enhancer has been published in Development today!
journals.biologists.com/dev/article/...

I am delighted that our paper exploring the impact of Neanderthal-derived variants on the activity of a disease-associated craniofacial enhancer has been published in Development today!
journals.biologists.com/dev/article/...
We propose to think along Marr's 3 levels: computational problem, algorithm, implementation
Check out our review:
arxiv.org/abs/2510.24536

We propose to think along Marr's 3 levels: computational problem, algorithm, implementation
Check out our review:
arxiv.org/abs/2510.24536
🗓️Register here for upcoming session and the entire series:
us06web.zoom.us/webinar/regi...

🗓️Register here for upcoming session and the entire series:
us06web.zoom.us/webinar/regi...
If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5

If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5

Huge thanks to @danielibrahim.bsky.social, @arnaudkr.bsky.social & @stemundi.bsky.social and fantastic people in their labs — what a journey! 🧪🔬
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

Huge thanks to @danielibrahim.bsky.social, @arnaudkr.bsky.social & @stemundi.bsky.social and fantastic people in their labs — what a journey! 🧪🔬
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you
Together with Christine Moene @cmoene.bsky.social, we explored what happens when you scramble the genome—revealing how Sox2’s position shapes enhancer activation.
📖 Read the full story here: www.science.org/doi/10.1126/...

Together with Christine Moene @cmoene.bsky.social, we explored what happens when you scramble the genome—revealing how Sox2’s position shapes enhancer activation.
📖 Read the full story here: www.science.org/doi/10.1126/...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/
Deadline July 15th.
www.mdc-berlin.de/career/jobs/...
Deadline July 15th.
www.mdc-berlin.de/career/jobs/...
www.fmi.ch/education-ca...
www.fmi.ch/education-ca...
Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro

Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro
www.science.org/doi/10.1126/...
A short clip describing the key results:
www.youtube.com/watch?v=pmvO...

www.science.org/doi/10.1126/...
A short clip describing the key results:
www.youtube.com/watch?v=pmvO...
www.nature.com/articles/s41...
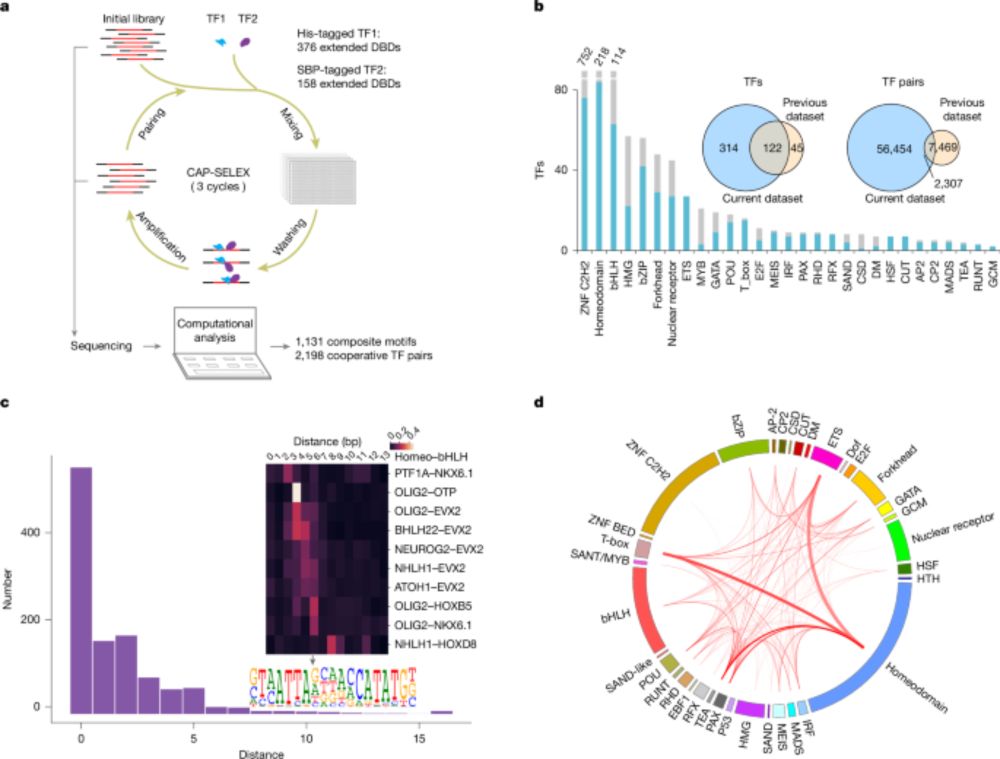
www.nature.com/articles/s41...
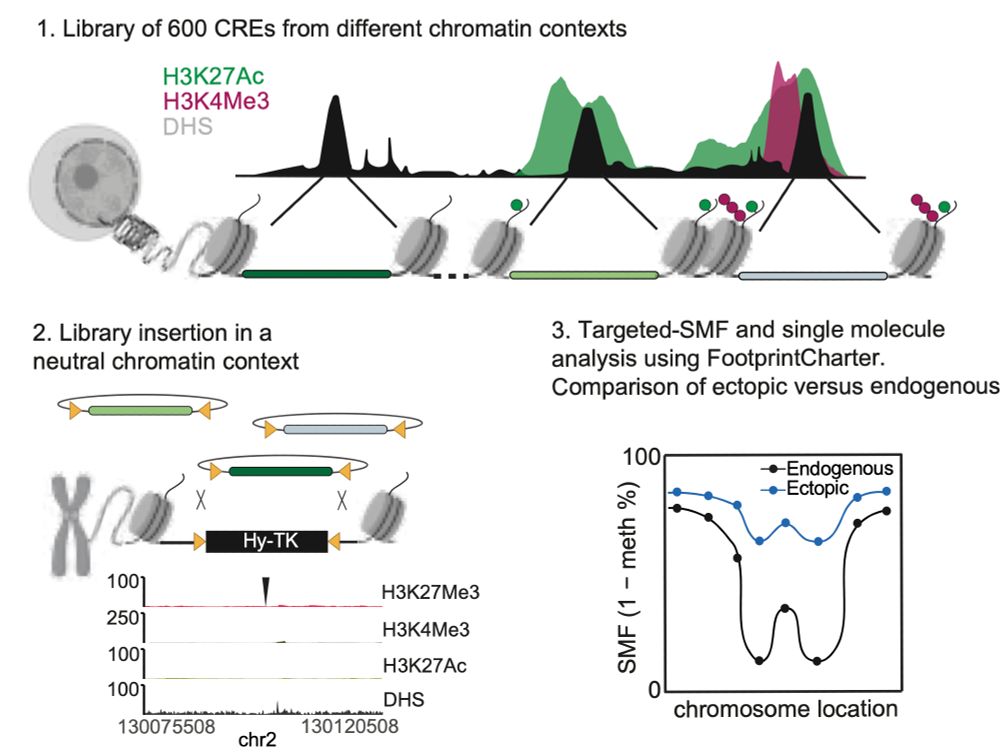
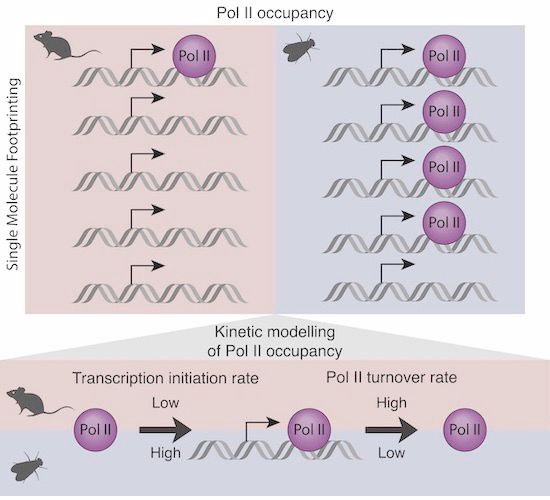
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

