
🔹 Support for ~170 bioinformatics tools
🔹 1 run every 3 seconds (25k+ daily runs)
🔹 From single contributor to 235 contributors
🔹 7000+ journal citations (~8-9 per day)
📚 Discover a timeline of innovation:
hubs.ly/Q03PsbTP0
🔹 Support for ~170 bioinformatics tools
🔹 1 run every 3 seconds (25k+ daily runs)
🔹 From single contributor to 235 contributors
🔹 7000+ journal citations (~8-9 per day)
📚 Discover a timeline of innovation:
hubs.ly/Q03PsbTP0
What began as a simple QC reporting tool has evolved into an essential resource used by thousands worldwide, becoming integral to the #bioinformatics ecosystm.
🔍 Discover MultiQC: hubs.la/Q03Bh0wM0

What began as a simple QC reporting tool has evolved into an essential resource used by thousands worldwide, becoming integral to the #bioinformatics ecosystm.
🔍 Discover MultiQC: hubs.la/Q03Bh0wM0
What started as a simple tool for QC reports has grown into something used by thousands of labs worldwide. It's become a foundational tool in the fabric of #bioinformatics and it's a joy to see it play a small part in global research
What started as a simple tool for QC reports has grown into something used by thousands of labs worldwide. It's become a foundational tool in the fabric of #bioinformatics and it's a joy to see it play a small part in global research
Tools you need:
FastQC: first defense
MultiQC: overview across samples
IGV: visualize coverage & peaks
PCA plots: always
Links:
www.bioinformatics.babraham.ac.uk/projects/fa...
multiqc.info/
software.broadinstitute.org/software/igv/
Tools you need:
FastQC: first defense
MultiQC: overview across samples
IGV: visualize coverage & peaks
PCA plots: always
Links:
www.bioinformatics.babraham.ac.uk/projects/fa...
multiqc.info/
software.broadinstitute.org/software/igv/
There are a handful of release highlights for this version, but perhaps the most exciting is that .gif, .webp and .tiff files are now supported for #CustomContent - you can finally put your favourite gifs into reports! Just name them *_mqc.gif

There are a handful of release highlights for this version, but perhaps the most exciting is that .gif, .webp and .tiff files are now supported for #CustomContent - you can finally put your favourite gifs into reports! Just name them *_mqc.gif
RNAseqqc github.com/getzlab/rna...
🔗 Check out multiQC for aggregated reports:multiqc.info/ multiqc.info/🔬 Learn to visualize data with IGV: software.broadinstitute.org/software/igv/
Start every analysis with rigorous QC! 🚀
RNAseqqc github.com/getzlab/rna...
🔗 Check out multiQC for aggregated reports:multiqc.info/ multiqc.info/🔬 Learn to visualize data with IGV: software.broadinstitute.org/software/igv/
Start every analysis with rigorous QC! 🚀
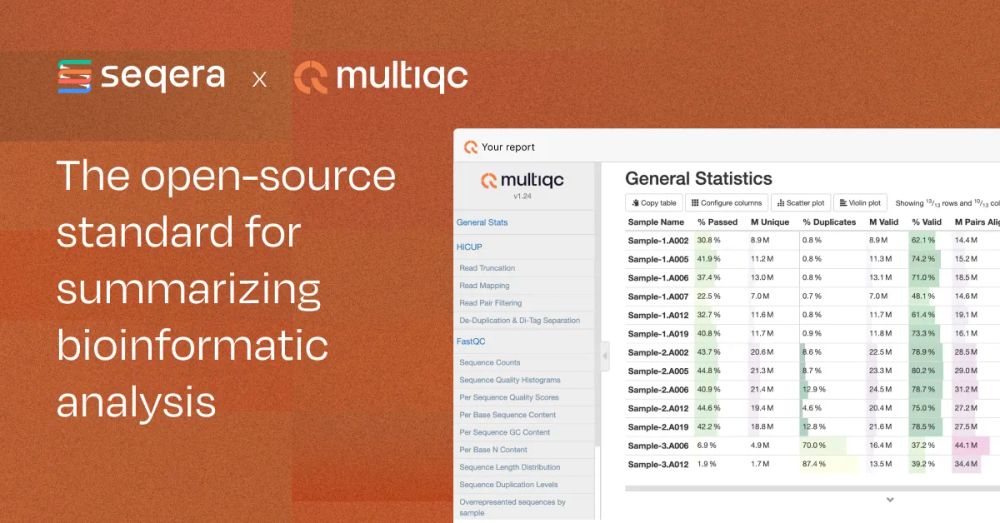
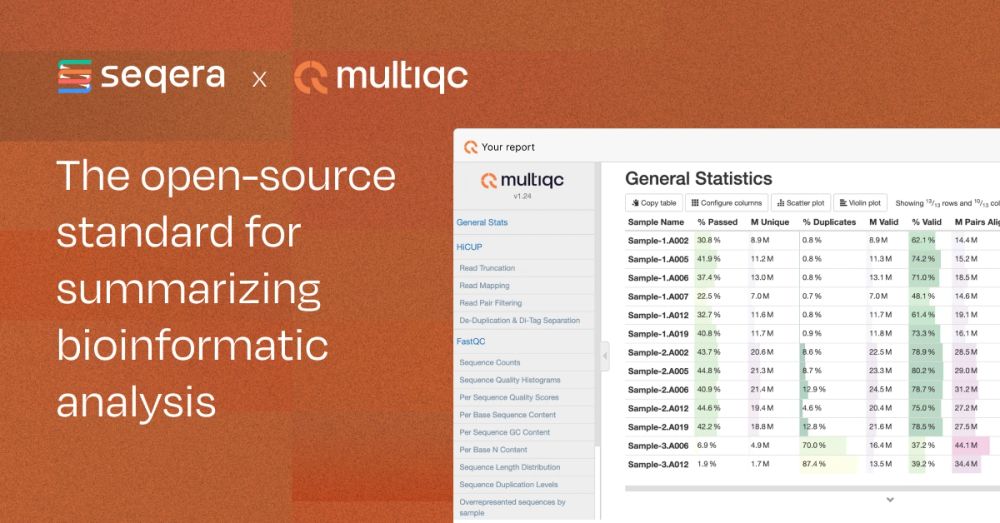
seqera.io/multiqc/
github.com/MultiQC/Mult...
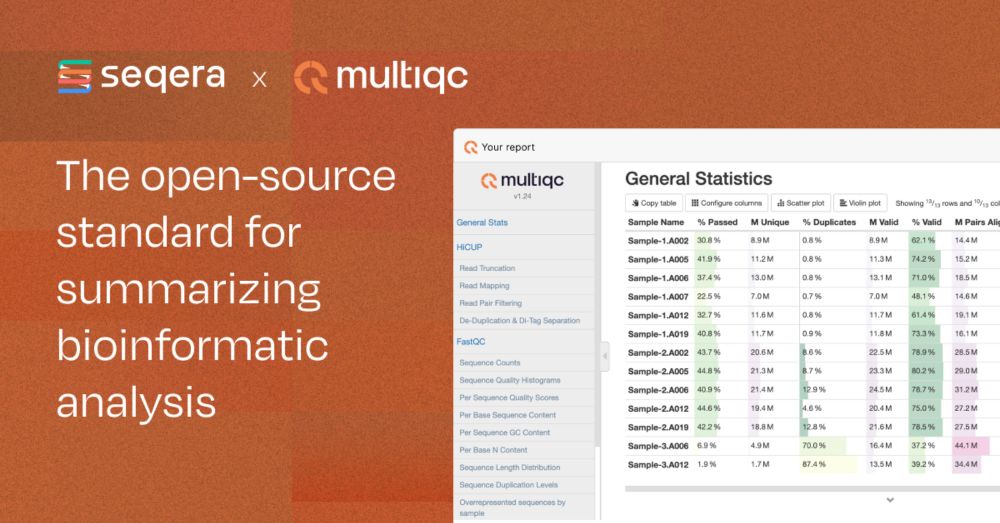
seqera.io/multiqc/
github.com/MultiQC/Mult...


Find out more: hubs.la/Q033Kjp30
Find out more: hubs.la/Q033Kjp30

=== ✨ AI summaries ✨ ===
LLMs are a natural next step for helping scientists understand large datasets. @multiqc.info now has built-in integration with @seqera.io AI as well as #OpenAI and #AnthropicAI 🤖
=== ✨ AI summaries ✨ ===
LLMs are a natural next step for helping scientists understand large datasets. @multiqc.info now has built-in integration with @seqera.io AI as well as #OpenAI and #AnthropicAI 🤖
- MosaiCatcher
- ataqv
- telseq
- Cell
- GTDB
- Checkm
- Checkm2
- Haplocheck
- mgikit
- Percolator
- MosaiCatcher
- ataqv
- telseq
- Cell
- GTDB
- Checkm
- Checkm2
- Haplocheck
- mgikit
- Percolator
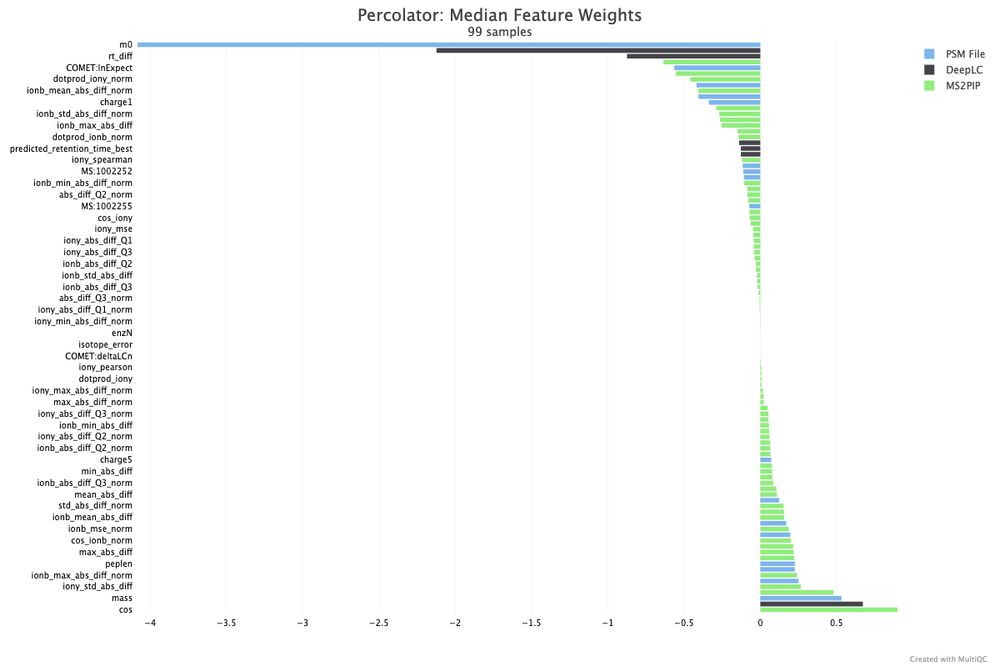
- 🆕 10 new tools supported
- 💫 12 modules updated
- 🛠️ Bug fixes, improvements and refactoring!
If you're quick, you might see a little christmassy easter egg!
github.com/MultiQC/Mult...
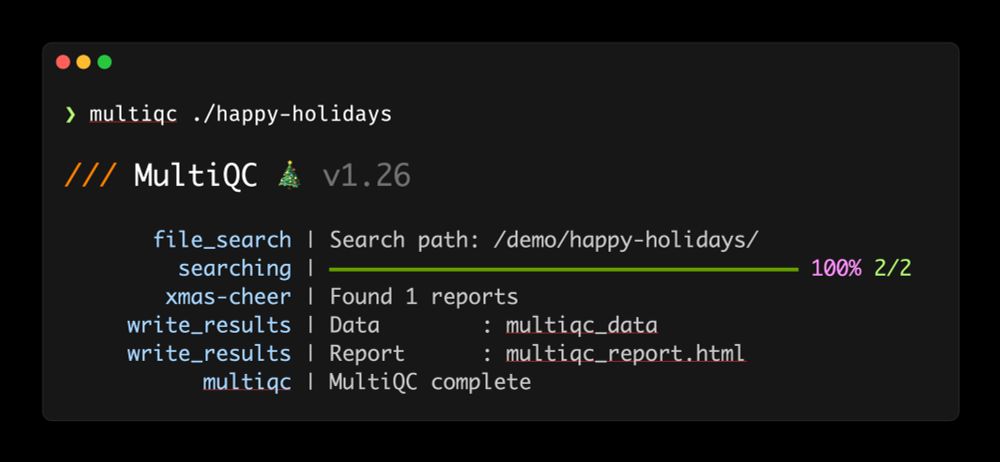
- 🆕 10 new tools supported
- 💫 12 modules updated
- 🛠️ Bug fixes, improvements and refactoring!
If you're quick, you might see a little christmassy easter egg!
github.com/MultiQC/Mult...
Catch errors early and save time on downstream analysis!
training.galaxyproject.org/training-mat...
Catch errors early and save time on downstream analysis!
training.galaxyproject.org/training-mat...
Every MultiQC config option can now be set via an env var, and YAML configs can interpolate env vars into their strings:
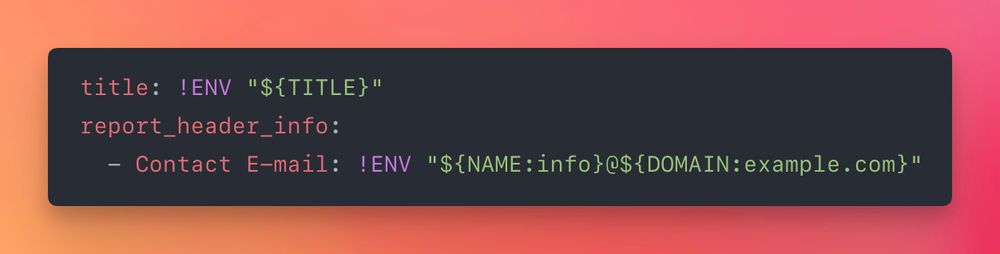
Every MultiQC config option can now be set via an env var, and YAML configs can interpolate env vars into their strings:
This patch release fixes a whole host of bugs, the most important one being for #bcl2fastq 🐛
There are also a host of small additions sneaking in for #ngsbits, #picard, #Nanostat, #Samtools and more - so well worth the update!
github.com/MultiQC/Mult...

This patch release fixes a whole host of bugs, the most important one being for #bcl2fastq 🐛
There are also a host of small additions sneaking in for #ngsbits, #picard, #Nanostat, #Samtools and more - so well worth the update!
github.com/MultiQC/Mult...

If you're new to @multiqc.info then check out seqera.io/multiqc/ to see what it's all about and have a go at running MultiQC in your browser!

If you're new to @multiqc.info then check out seqera.io/multiqc/ to see what it's all about and have a go at running MultiQC in your browser!

