

Here's to the next decade of making #bioinformatics QC beautiful! 🧬
Here's to the next decade of making #bioinformatics QC beautiful! 🧬
Tools you need:
FastQC: first defense
MultiQC: overview across samples
IGV: visualize coverage & peaks
PCA plots: always
Links:
www.bioinformatics.babraham.ac.uk/projects/fa...
multiqc.info/
software.broadinstitute.org/software/igv/
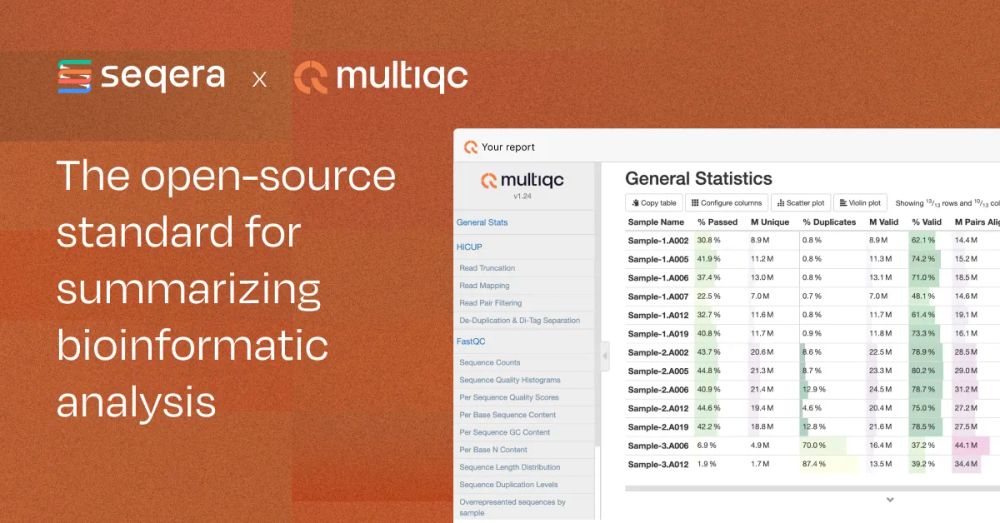
Tools you need:
FastQC: first defense
MultiQC: overview across samples
IGV: visualize coverage & peaks
PCA plots: always
Links:
www.bioinformatics.babraham.ac.uk/projects/fa...
multiqc.info/
software.broadinstitute.org/software/igv/
See the full changelog here: github.com/MultiQC/Mult...

See the full changelog here: github.com/MultiQC/Mult...
See the docs to get started: docs.seqera.io/multiqc/gett...

See the docs to get started: docs.seqera.io/multiqc/gett...
This has been added to schemastore.org so that config files in #VSCode & other IDEs automatically get integrated error checking as you type, see docs.seqera.io/multiqc/gett...
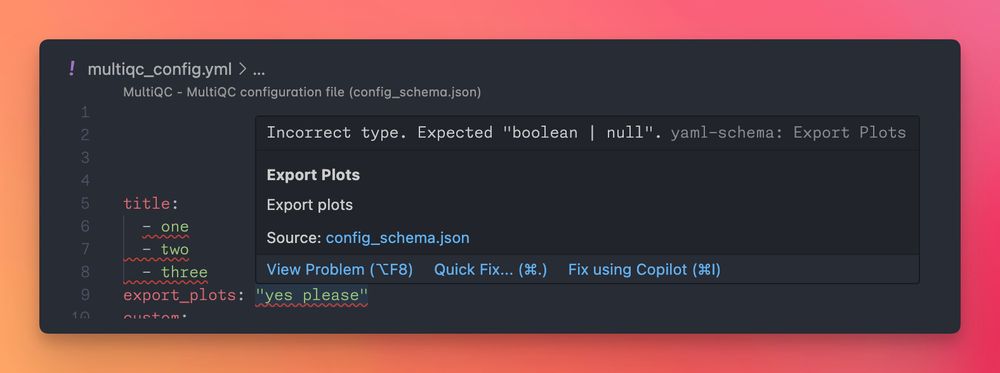
This has been added to schemastore.org so that config files in #VSCode & other IDEs automatically get integrated error checking as you type, see docs.seqera.io/multiqc/gett...
RNAseqqc github.com/getzlab/rna...
🔗 Check out multiQC for aggregated reports:multiqc.info/ multiqc.info/🔬 Learn to visualize data with IGV: software.broadinstitute.org/software/igv/
Start every analysis with rigorous QC! 🚀
RNAseqqc github.com/getzlab/rna...
🔗 Check out multiQC for aggregated reports:multiqc.info/ multiqc.info/🔬 Learn to visualize data with IGV: software.broadinstitute.org/software/igv/
Start every analysis with rigorous QC! 🚀

github.com/MultiQC/Mult...

github.com/MultiQC/Mult...
Reports also have "Copy prompt" options to paste into whatever LLM you like.
Read more in the blog post: hubs.ly/Q033KvM10

Reports also have "Copy prompt" options to paste into whatever LLM you like.
Read more in the blog post: hubs.ly/Q033KvM10
This can be done at report-generation time, with the summary is part of the HTML. But also *interactively* on-demand when reading the report.
This can be done at report-generation time, with the summary is part of the HTML. But also *interactively* on-demand when reading the report.
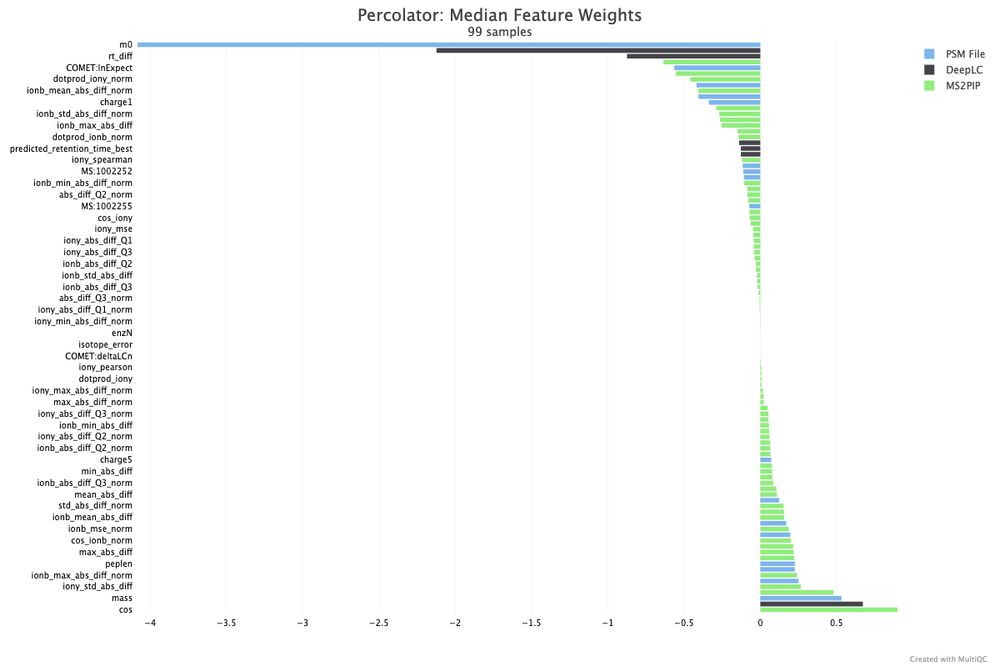
github.com/MultiQC/Mult...

github.com/MultiQC/Mult...
- GATK BQSR
- Pychopper
- fastp
- RNA-SeqC
- phantompeakqualtools
- QualiMap RNASeq
- QualiMap BamQC
- DRAGEN
- Interop
- Dedup
- Bclconvert
- Nanostat
- GATK BQSR
- Pychopper
- fastp
- RNA-SeqC
- phantompeakqualtools
- QualiMap RNASeq
- QualiMap BamQC
- DRAGEN
- Interop
- Dedup
- Bclconvert
- Nanostat

