Please see the changelog: https://github.com/nf-core/epitopeprediction/releases/tag/3.1.0

Please see the changelog: https://github.com/nf-core/epitopeprediction/releases/tag/3.1.0
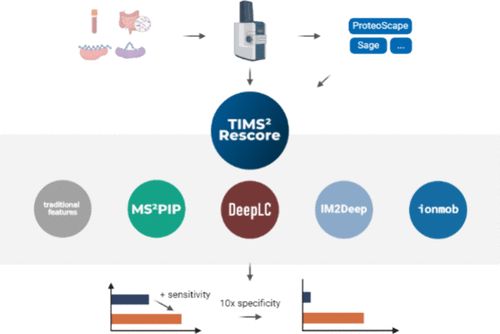
An open-source @nf-co.re workflow using OpenMS, DeepLC & MS²PIP boosts peptide ID & sensitivity. Scalable, reproducible & ready for large-scale immunopeptidomics.
👉 rdcu.be/eHFTZ
An open-source @nf-co.re workflow using OpenMS, DeepLC & MS²PIP boosts peptide ID & sensitivity. Scalable, reproducible & ready for large-scale immunopeptidomics.
👉 rdcu.be/eHFTZ

Please see the changelog: https://github.com/nf-core/hlatyping/releases/tag/2.1.0
Please see the changelog: https://github.com/nf-core/hlatyping/releases/tag/2.1.0
Answer our survey for improving nf-core/airrflow, a Nextflow pipeline to analyze bulk and single-cell AIRRseq data (doi.org/10.1371/jour...). We’re interested in your opinion on useful new features!
yalesurvey.ca1.qualtrics.com/jfe/form/SV_...
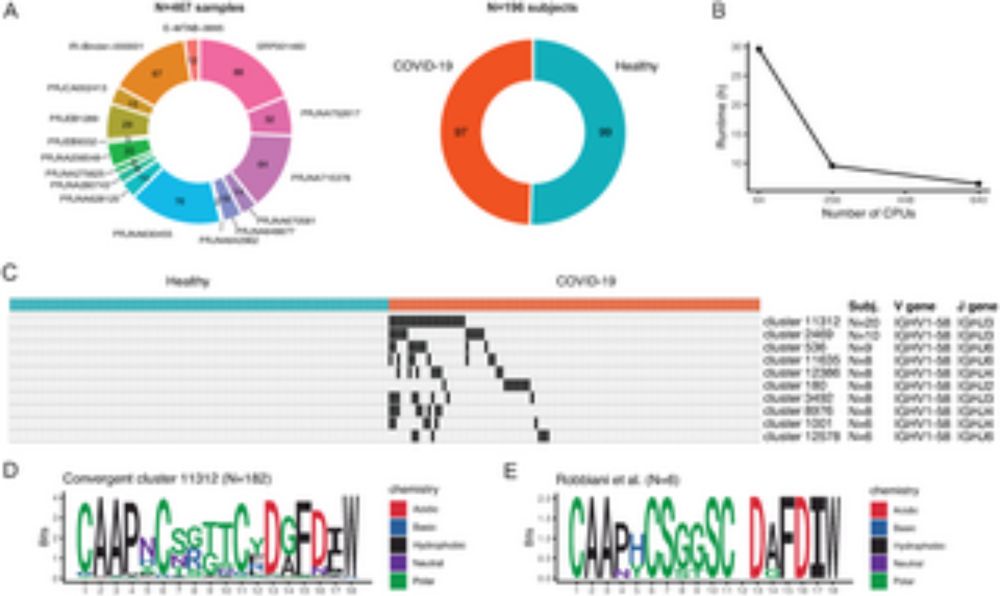
Answer our survey for improving nf-core/airrflow, a Nextflow pipeline to analyze bulk and single-cell AIRRseq data (doi.org/10.1371/jour...). We’re interested in your opinion on useful new features!
yalesurvey.ca1.qualtrics.com/jfe/form/SV_...
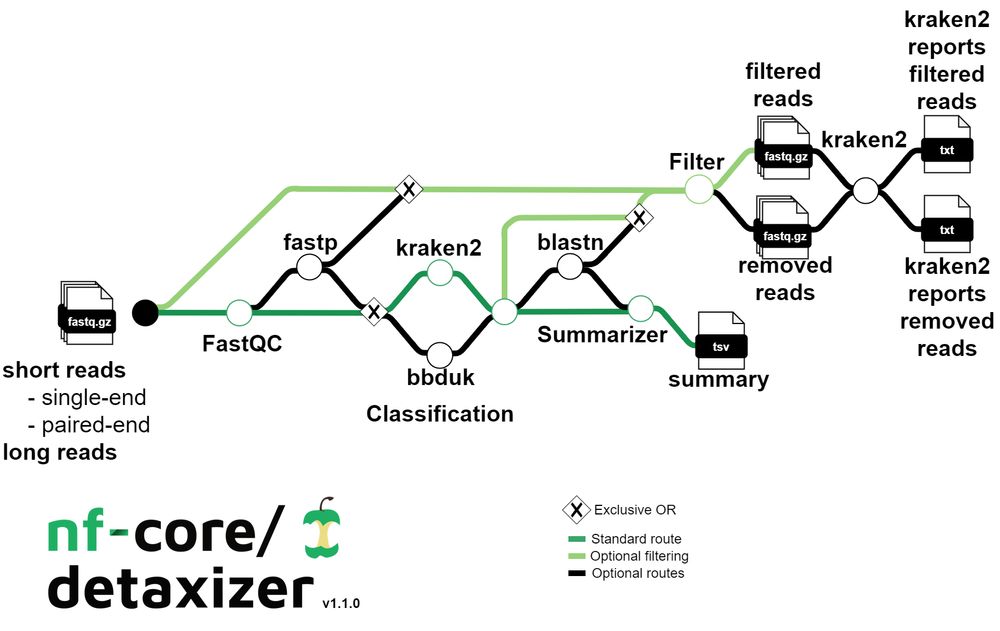
nf-co.re/events/2025/...

nf-co.re/events/2025/...
---
#proteomics #prot-paper
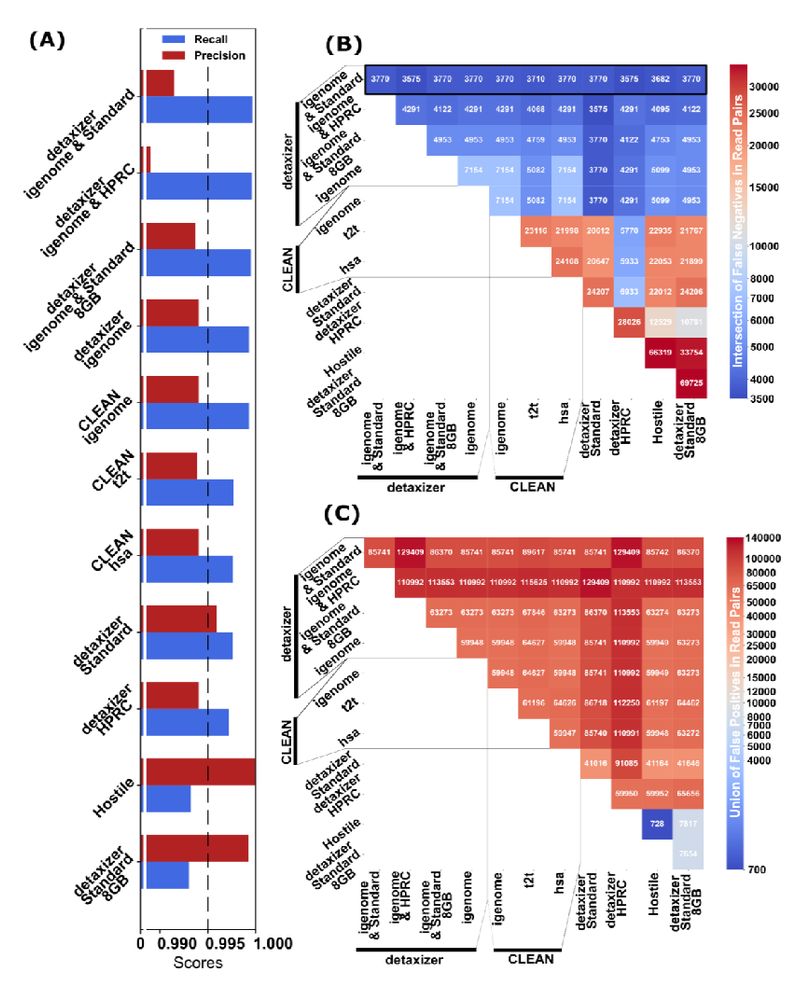
---
#proteomics #prot-paper
#EuBIC2025




#EuBIC2025
Specifically, for people in the quantitative sciences, I recommend:
- Have a well-tended ORCID
- GitHub/GitLab repository with some of your research outputs
- Personal webpage (e.g. quarto) on a static hosting service such as GitHub/GitLab Pages, Netlify, Posit Connect, ...
LinkedIn is a closed, for-profit service. If you don't have an account, you can't see someone else's profile.
I do not have LinkedIn. I can't see your profile.
If you are job/internship/PhD hunting I recommend having an open online presence!
pubs.acs.org/doi/abs/10.1...
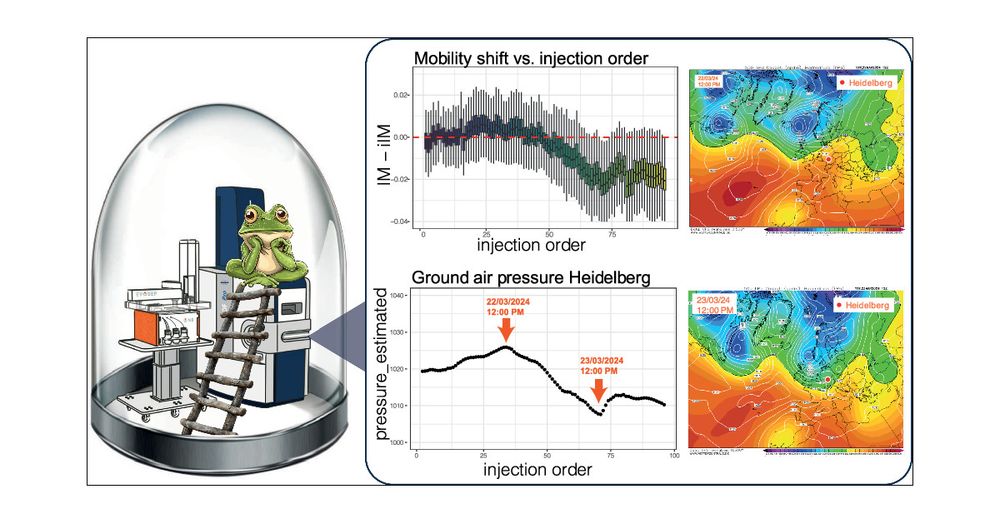
pubs.acs.org/doi/abs/10.1...
Next: Follow-up trial with improved adjuvants & BTK inhibitors underway. Stay tuned! #CancerResearch #Immunopeptidomics #Immunotherapy
🔗 doi.org/10.3389/fimm...
Next: Follow-up trial with improved adjuvants & BTK inhibitors underway. Stay tuned! #CancerResearch #Immunopeptidomics #Immunotherapy
🔗 doi.org/10.3389/fimm...
---
#proteomics #prot-preprint
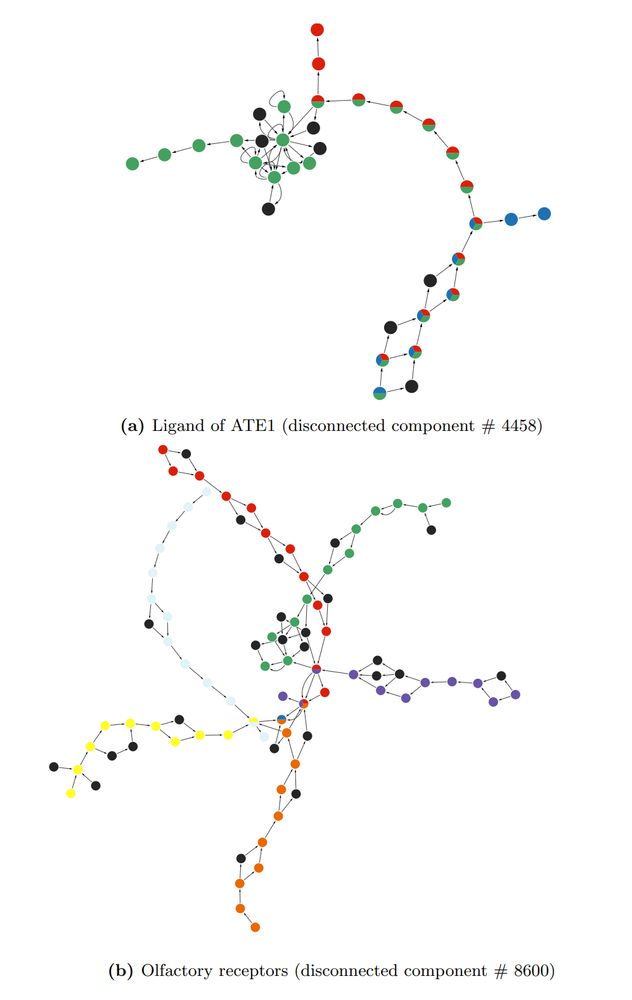
---
#proteomics #prot-preprint
A big thanks to Louise Buur and Viktoria Dorfer for the great collaboration!
Open access full text: biblio.ugent.be/publication/...
Project: github.com/compomics/ms...
A big thanks to Louise Buur and Viktoria Dorfer for the great collaboration!
Open access full text: biblio.ugent.be/publication/...
Project: github.com/compomics/ms...
Please see the changelog: https://github.com/nf-core/epitopeprediction/releases/tag/2.3.0
Please see the changelog: https://github.com/nf-core/epitopeprediction/releases/tag/2.3.0

