

Tons of open slots, invited and selected speakers all get 20 mins ❤️
#EESNucleus
Tons of open slots, invited and selected speakers all get 20 mins ❤️
#EESNucleus
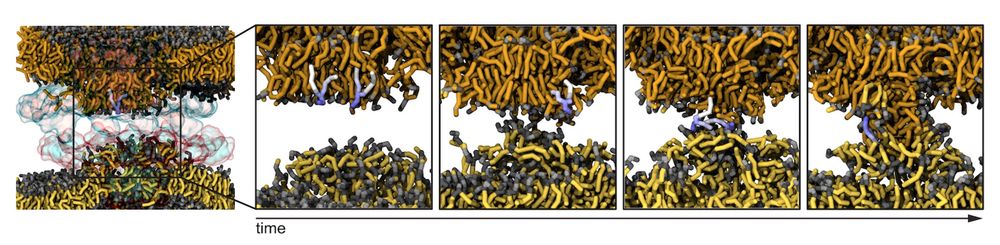
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
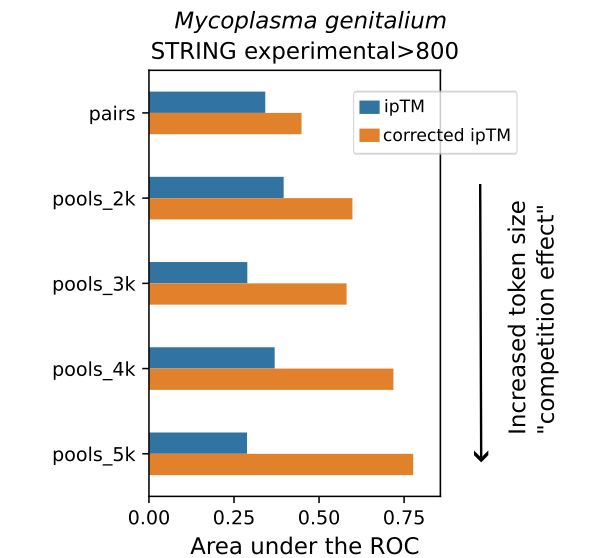
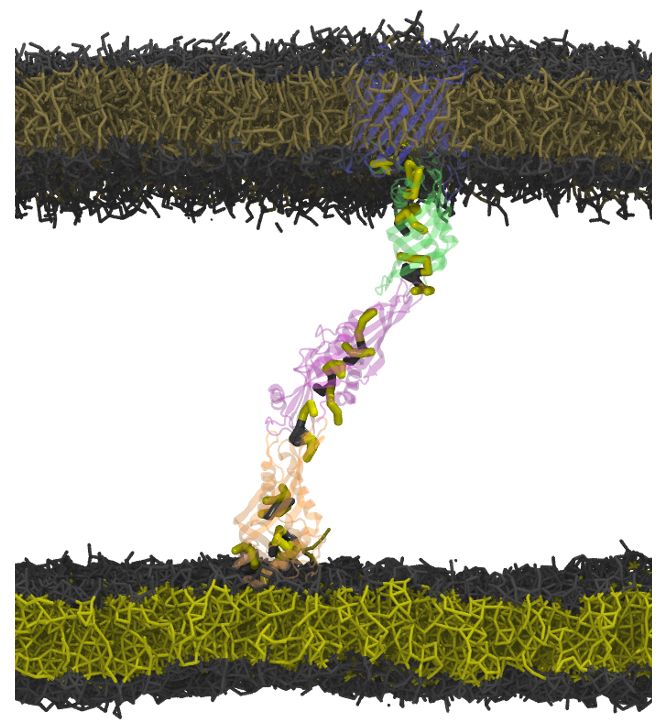
pubs.acs.org/doi/full/10....

pubs.acs.org/doi/full/10....
buff.ly/4b3E0xy

buff.ly/4b3E0xy
rupress.org/jcb/article/...
rupress.org/jcb/article/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪
www.nature.com/articles/s41...
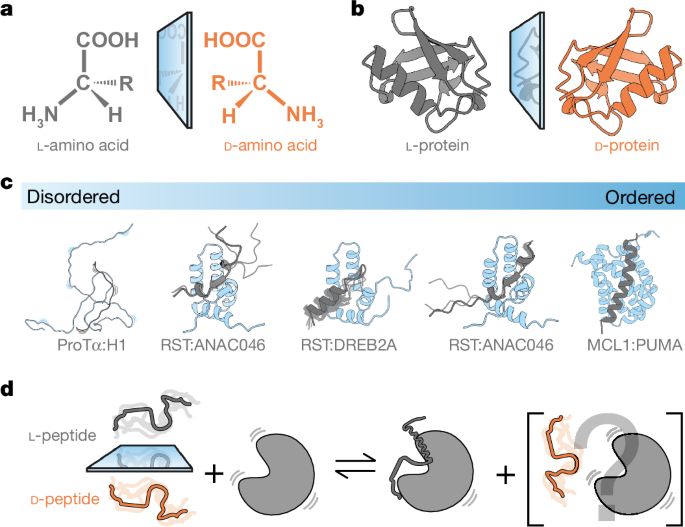
www.nature.com/articles/s41...

Identifying Sequence Effects on Chain Dimensions of Disordered Proteins by Integrating Experiments and Simulations.
pubs.acs.org/doi/10.1021/...
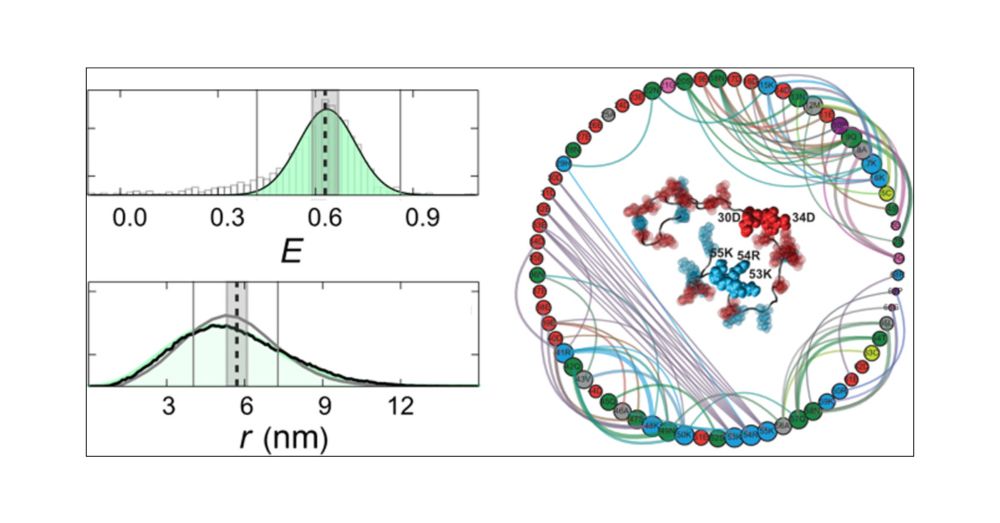
Identifying Sequence Effects on Chain Dimensions of Disordered Proteins by Integrating Experiments and Simulations.
pubs.acs.org/doi/10.1021/...


Statistical mechanics effectively says: why bother with all those complex trajectories? Just go ahead and replace them with truly random motion.
Statistical mechanics effectively says: why bother with all those complex trajectories? Just go ahead and replace them with truly random motion.


