Milot Mirdita
@milot.bsky.social
Open source methods for #metagenomics with @martinsteinegger.bsky.social Seoul National University 🇰🇷 | former Söding Lab MPI-NAT 🇩🇪 | http://mstdn.science/@milotmirdita
Reposted by Milot Mirdita
I want to spell this out in case the implications aren't clear:
This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.
This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.
On Oct 1, 2025, GISAID informed us that they had ended updates to the flat file of SARS-CoV-2 genomic sequences and associated metadata that we had used to update Nextstrain analyses since Feb 2020. GISAID's stated rationale was that their "resources are limited". 1/5

November 7, 2025 at 2:41 PM
I want to spell this out in case the implications aren't clear:
This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.
This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.
Reposted by Milot Mirdita
OpenFold3-preview (OF3p) is out: a sneak peek of our AF3-based structure prediction model. Our aim for OF3 is full AF3-parity for every modality. We now believe we have a clear path towards this goal and are releasing OF3p to enable building in the OF3 ecosystem. More👇
October 28, 2025 at 6:30 PM
OpenFold3-preview (OF3p) is out: a sneak peek of our AF3-based structure prediction model. Our aim for OF3 is full AF3-parity for every modality. We now believe we have a clear path towards this goal and are releasing OF3p to enable building in the OF3 ecosystem. More👇
Reposted by Milot Mirdita
DIAMOND v2.1.15 now supports all taxonomy features for BLAST databases, and support for using BLAST databases has also been added to the Bioconda version github.com/bbuchfink/di...

GitHub - bbuchfink/diamond: Accelerated BLAST compatible local sequence aligner.
Accelerated BLAST compatible local sequence aligner. - bbuchfink/diamond
github.com
October 28, 2025 at 4:45 PM
DIAMOND v2.1.15 now supports all taxonomy features for BLAST databases, and support for using BLAST databases has also been added to the Bioconda version github.com/bbuchfink/di...
Reposted by Milot Mirdita
Our new preprint is out. Our group performed a comprehensive protein–protein complex prediction within 2,437 biosynthetic gene clusters. We predicted a total of 487,828 complexes for known BGCs, identifying 15,438 heteromeric interactions with an ipTM ≥ 0.6. (2/3)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Predicting protein complexes in biosynthetic gene clusters
Biosynthetic gene clusters (BGCs) are contiguous genomic regions that encode diverse, non-homologous proteins required for the production of specific natural products. Their genetic diversity underlie...
www.biorxiv.org
October 28, 2025 at 5:58 AM
Our new preprint is out. Our group performed a comprehensive protein–protein complex prediction within 2,437 biosynthetic gene clusters. We predicted a total of 487,828 complexes for known BGCs, identifying 15,438 heteromeric interactions with an ipTM ≥ 0.6. (2/3)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Milot Mirdita
October 28, 2025 at 3:34 AM
Reposted by Milot Mirdita
Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project is collaborating with a broad network of leading wetlabs that test BoltzGen at an unprecedented scale, showing success on many novel targets and pushing the model to its limits!

October 26, 2025 at 10:40 PM
Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project is collaborating with a broad network of leading wetlabs that test BoltzGen at an unprecedented scale, showing success on many novel targets and pushing the model to its limits!
Reposted by Milot Mirdita
We train machine learning models on millions of proteins. But when it comes to making predictions, do we need them to understand all proteins at once? Often, we need an accurate model for the specific protein we are studying or designing. We address this with ProteinTTT arxiv.org/abs/2411.02109 1/🧵
October 23, 2025 at 1:08 PM
We train machine learning models on millions of proteins. But when it comes to making predictions, do we need them to understand all proteins at once? Often, we need an accurate model for the specific protein we are studying or designing. We address this with ProteinTTT arxiv.org/abs/2411.02109 1/🧵
Reposted by Milot Mirdita
End-to-end protein design in the browser through evedesign. Generate and interactively explore designs in 2D/3D and export them as codon-optimized DNA. The underlying open source framework (released soon) is build to easily add new methods, more on that soon.
🌐 evedesign.bio
🌐 evedesign.bio
October 22, 2025 at 2:30 PM
End-to-end protein design in the browser through evedesign. Generate and interactively explore designs in 2D/3D and export them as codon-optimized DNA. The underlying open source framework (released soon) is build to easily add new methods, more on that soon.
🌐 evedesign.bio
🌐 evedesign.bio
Reposted by Milot Mirdita
Announcing our new protein design server evedesign.bio:
• End-to-end protein design for everyone!
• Analyze your generated library interactively and on 3D structures
• Export codon-optimized DNA sequences for experimental testing.
• End-to-end protein design for everyone!
• Analyze your generated library interactively and on 3D structures
• Export codon-optimized DNA sequences for experimental testing.
October 22, 2025 at 2:17 PM
Announcing our new protein design server evedesign.bio:
• End-to-end protein design for everyone!
• Analyze your generated library interactively and on 3D structures
• Export codon-optimized DNA sequences for experimental testing.
• End-to-end protein design for everyone!
• Analyze your generated library interactively and on 3D structures
• Export codon-optimized DNA sequences for experimental testing.
Reposted by Milot Mirdita
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

October 20, 2025 at 11:26 AM
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Reposted by Milot Mirdita
Our Metabuli taxonomic classifier is now available as an app compatible with Windows, Linux, and macOS. It features an interactive visualization developed by @sunjaelee.bsky.social, along with improvements to Metabuli by @jbeom.bsky.social.
📄 academic.oup.com/bioinformati...
📄 academic.oup.com/bioinformati...
October 16, 2025 at 1:01 PM
Our Metabuli taxonomic classifier is now available as an app compatible with Windows, Linux, and macOS. It features an interactive visualization developed by @sunjaelee.bsky.social, along with improvements to Metabuli by @jbeom.bsky.social.
📄 academic.oup.com/bioinformati...
📄 academic.oup.com/bioinformati...
Reposted by Milot Mirdita
Easy and interactive taxonomic profiling with Metabuli App.
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
October 16, 2025 at 7:29 AM
Easy and interactive taxonomic profiling with Metabuli App.
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
Reposted by Milot Mirdita
Easy and interactive taxonomic profiling with Metabuli App academic.oup.com/bioinformati... 🧬🖥️🧪 github.com/steineggerla...

October 14, 2025 at 5:55 PM
Easy and interactive taxonomic profiling with Metabuli App academic.oup.com/bioinformati... 🧬🖥️🧪 github.com/steineggerla...
AFDB v6 is already available in Foldseek!
🚀 search.foldseek.com
foldseek databases Alphafold/UniProt50 afdb50 tmp
foldseek easy-search query.cif afdb50 res.m8 tmp --cluster-search 0/1 (Reps only / all 241M)
Thanks @ebi.embl.org and DeepMind for working with us to make it available to everyone.
🚀 search.foldseek.com
foldseek databases Alphafold/UniProt50 afdb50 tmp
foldseek easy-search query.cif afdb50 res.m8 tmp --cluster-search 0/1 (Reps only / all 241M)
Thanks @ebi.embl.org and DeepMind for working with us to make it available to everyone.
We’re renewing our collaboration with Google DeepMind!
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social

October 7, 2025 at 2:33 PM
AFDB v6 is already available in Foldseek!
🚀 search.foldseek.com
foldseek databases Alphafold/UniProt50 afdb50 tmp
foldseek easy-search query.cif afdb50 res.m8 tmp --cluster-search 0/1 (Reps only / all 241M)
Thanks @ebi.embl.org and DeepMind for working with us to make it available to everyone.
🚀 search.foldseek.com
foldseek databases Alphafold/UniProt50 afdb50 tmp
foldseek easy-search query.cif afdb50 res.m8 tmp --cluster-search 0/1 (Reps only / all 241M)
Thanks @ebi.embl.org and DeepMind for working with us to make it available to everyone.
Reposted by Milot Mirdita
We’re renewing our collaboration with Google DeepMind!
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social

October 7, 2025 at 2:03 PM
We’re renewing our collaboration with Google DeepMind!
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social
Reposted by Milot Mirdita
New exciting resource published!! The Viro3D paper is out, describing our comprehensive database of predicted virus protein structures! 💻🧬
Work with @ulad-litvin.bsky.social , @grovearmada.bsky.social , Alex Jack, @bljog.bsky.social , @davidlrobertson.bsky.social
www.embopress.org/doi/full/10....
Work with @ulad-litvin.bsky.social , @grovearmada.bsky.social , Alex Jack, @bljog.bsky.social , @davidlrobertson.bsky.social
www.embopress.org/doi/full/10....

Viro3D: a comprehensive database of virus protein structure predictions | Molecular Systems Biology
imageimageViro3D provides proteome-level, high confidence AI-protein structure predictions for
>4,400 viruses, allowing mapping of form and function across the human and animal
virosphere. Viro3D i...
www.embopress.org
September 28, 2025 at 5:39 AM
New exciting resource published!! The Viro3D paper is out, describing our comprehensive database of predicted virus protein structures! 💻🧬
Work with @ulad-litvin.bsky.social , @grovearmada.bsky.social , Alex Jack, @bljog.bsky.social , @davidlrobertson.bsky.social
www.embopress.org/doi/full/10....
Work with @ulad-litvin.bsky.social , @grovearmada.bsky.social , Alex Jack, @bljog.bsky.social , @davidlrobertson.bsky.social
www.embopress.org/doi/full/10....
Reposted by Milot Mirdita
#RECOMB2026 will be in Thessaloniki, Greece on May 26-29, 2026. Satellites on May 24-25. Save the date!
Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!
Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!

September 26, 2025 at 3:03 PM
#RECOMB2026 will be in Thessaloniki, Greece on May 26-29, 2026. Satellites on May 24-25. Save the date!
Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!
Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!
Reposted by Milot Mirdita
HHMI adopts Plan U journals.plos.org/plosbiology/...

September 24, 2025 at 6:08 PM
HHMI adopts Plan U journals.plos.org/plosbiology/...
Reposted by Milot Mirdita
Delighted to finally announce a preprint describing the Q100 project! “A complete diploid human genome benchmark for personalized genomics” For which we finished HG002 to near-perfect accuracy: www.biorxiv.org/content/10.1... 🧵[1/14]

A complete diploid human genome benchmark for personalized genomics
Human genome resequencing typically involves mapping reads to a reference genome to call variants; however, this approach suffers from both technical and reference biases, leaving many duplicated and ...
www.biorxiv.org
September 22, 2025 at 5:01 PM
Delighted to finally announce a preprint describing the Q100 project! “A complete diploid human genome benchmark for personalized genomics” For which we finished HG002 to near-perfect accuracy: www.biorxiv.org/content/10.1... 🧵[1/14]
Reposted by Milot Mirdita
GPU-accelerated MMseqs2 offers tremendous speedup for homology retrieval, protein structure prediction with ColabFold, and protein structure search with Foldseek. @martinsteinegger.bsky.social @milot.bsky.social @machine.learning.bio
www.nature.com/articles/s41...
www.nature.com/articles/s41...
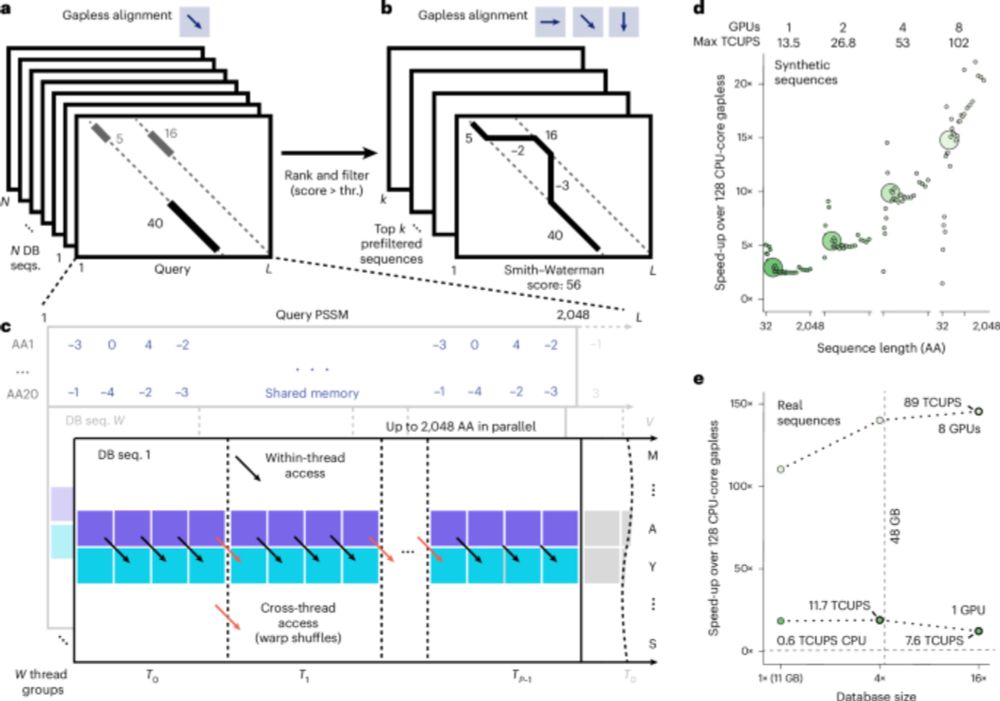
GPU-accelerated homology search with MMseqs2 - Nature Methods
Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure predictio...
www.nature.com
September 18, 2025 at 8:09 PM
GPU-accelerated MMseqs2 offers tremendous speedup for homology retrieval, protein structure prediction with ColabFold, and protein structure search with Foldseek. @martinsteinegger.bsky.social @milot.bsky.social @machine.learning.bio
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Milot Mirdita
MMseqs2-GPU sets new standards in single query search speed, allows near instant search of big databases, scales to multiple GPUs and is fast beyond VRAM. It enables ColabFold MSA generation in seconds and sub-second Foldseek search against AFDB50. 1/n
📄 www.nature.com/articles/s41...
💿 mmseqs.com
📄 www.nature.com/articles/s41...
💿 mmseqs.com

GPU-accelerated homology search with MMseqs2 - Nature Methods
Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure predictio...
www.nature.com
September 21, 2025 at 8:06 AM
MMseqs2-GPU sets new standards in single query search speed, allows near instant search of big databases, scales to multiple GPUs and is fast beyond VRAM. It enables ColabFold MSA generation in seconds and sub-second Foldseek search against AFDB50. 1/n
📄 www.nature.com/articles/s41...
💿 mmseqs.com
📄 www.nature.com/articles/s41...
💿 mmseqs.com
Reposted by Milot Mirdita
Happy to see that Spacedust is now published on Nature Methods!
It combines sensitive Foldseek structure search and conserved neighborhood detection to discover functionally-associated gene clusters in prokaryotic & viral genomes.
1/6🧵
It combines sensitive Foldseek structure search and conserved neighborhood detection to discover functionally-associated gene clusters in prokaryotic & viral genomes.
1/6🧵
Spacedust: a tool for de novo identification of conserved gene clusters from metagenomic data.
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...

De novo discovery of conserved gene clusters in microbial genomes with Spacedust - Nature Methods
This work presents Spacedust, a tool for de novo identification of conserved gene clusters from metagenomic data.
www.nature.com
September 16, 2025 at 8:26 PM
Happy to see that Spacedust is now published on Nature Methods!
It combines sensitive Foldseek structure search and conserved neighborhood detection to discover functionally-associated gene clusters in prokaryotic & viral genomes.
1/6🧵
It combines sensitive Foldseek structure search and conserved neighborhood detection to discover functionally-associated gene clusters in prokaryotic & viral genomes.
1/6🧵
Reposted by Milot Mirdita
Preprint:
Highly efficient protein structure prediction on NVIDIA RTX Blackwell and Grace-Hopper
nvda.ws/4n4xzz9
Visit the NVIDIA Digital Biology Labs website to find more information like this:
t.co/R9ufEZrGEA
Highly efficient protein structure prediction on NVIDIA RTX Blackwell and Grace-Hopper
nvda.ws/4n4xzz9
Visit the NVIDIA Digital Biology Labs website to find more information like this:
t.co/R9ufEZrGEA
nvda.ws
September 16, 2025 at 1:59 PM
Preprint:
Highly efficient protein structure prediction on NVIDIA RTX Blackwell and Grace-Hopper
nvda.ws/4n4xzz9
Visit the NVIDIA Digital Biology Labs website to find more information like this:
t.co/R9ufEZrGEA
Highly efficient protein structure prediction on NVIDIA RTX Blackwell and Grace-Hopper
nvda.ws/4n4xzz9
Visit the NVIDIA Digital Biology Labs website to find more information like this:
t.co/R9ufEZrGEA
Reposted by Milot Mirdita
Spacedust: a tool for de novo identification of conserved gene clusters from metagenomic data.
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...

De novo discovery of conserved gene clusters in microbial genomes with Spacedust - Nature Methods
This work presents Spacedust, a tool for de novo identification of conserved gene clusters from metagenomic data.
www.nature.com
September 16, 2025 at 1:25 PM
Spacedust: a tool for de novo identification of conserved gene clusters from metagenomic data.
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
Reposted by Milot Mirdita


