
Michaela Novotná
@michaelanovotna.bsky.social
PhD student in @balikovalab.bsky.social at @czechacademy.bsky.social of Sciences, BIOCEV, @charlesuni.cuni.cz in Prague
💊 antibiotics 💎 ribosome
🧫 resistance 🧬 ABC-Fs
🦠 staph ❄️ cryo-em
@mbuavcr.bsky.social
@sciencecharles.bsky.social
💊 antibiotics 💎 ribosome
🧫 resistance 🧬 ABC-Fs
🦠 staph ❄️ cryo-em
@mbuavcr.bsky.social
@sciencecharles.bsky.social
Reposted by Michaela Novotná
`Mix-it-up': accessible time-resolved cryo-EM on the millisecond timescale pubmed.ncbi.nlm.nih.gov/41140236/ #cryoem
October 29, 2025 at 1:35 AM
`Mix-it-up': accessible time-resolved cryo-EM on the millisecond timescale pubmed.ncbi.nlm.nih.gov/41140236/ #cryoem
Reposted by Michaela Novotná
Mapping rRNA, tRNA, and mRNA modifications in ribosomes at high resolution: Trends in Biochemical Sciences www.cell.com/trends/bioch...

Mapping rRNA, tRNA, and mRNA modifications in ribosomes at high resolution
Chemical modifications of rRNA, tRNA, and mRNA play key roles in protein synthesis
by affecting the structure of these RNAs, by modulating decoding, and by influencing
ribosomal efficiency. Recent adv...
www.cell.com
October 24, 2025 at 8:28 AM
Mapping rRNA, tRNA, and mRNA modifications in ribosomes at high resolution: Trends in Biochemical Sciences www.cell.com/trends/bioch...
Reposted by Michaela Novotná
Reposted by Michaela Novotná
Check out our preprint! With new molecular mechanisms, 140 subtomogram averages, and ~600 annotated cells under different conditions, we @embl.org were able to describe bacterial populations with in-cell #cryoET. And there’s a surprise at the end 🕵️
www.biorxiv.org/content/10.1...
#teamtomo
www.biorxiv.org/content/10.1...
#teamtomo

October 15, 2025 at 6:26 AM
Check out our preprint! With new molecular mechanisms, 140 subtomogram averages, and ~600 annotated cells under different conditions, we @embl.org were able to describe bacterial populations with in-cell #cryoET. And there’s a surprise at the end 🕵️
www.biorxiv.org/content/10.1...
#teamtomo
www.biorxiv.org/content/10.1...
#teamtomo
Reposted by Michaela Novotná
Our work on ribosome hibernation in archaea is out!
We identified Hib, a new hibernation factor broadly distributed across archaea.
Check out the preprint 👉 biorxiv.org/content/10.1101/2025.10.11.676729v1
(1/🧵)
We identified Hib, a new hibernation factor broadly distributed across archaea.
Check out the preprint 👉 biorxiv.org/content/10.1101/2025.10.11.676729v1
(1/🧵)
October 13, 2025 at 4:26 PM
Our work on ribosome hibernation in archaea is out!
We identified Hib, a new hibernation factor broadly distributed across archaea.
Check out the preprint 👉 biorxiv.org/content/10.1101/2025.10.11.676729v1
(1/🧵)
We identified Hib, a new hibernation factor broadly distributed across archaea.
Check out the preprint 👉 biorxiv.org/content/10.1101/2025.10.11.676729v1
(1/🧵)
Reposted by Michaela Novotná
Dvě vědkyně z @mbuavcr.bsky.social - Natalia Slonská a Lucie Najmanová, se zúčastnily soutěže Věda fotogenická, kterou pořádá @akademievedcr.bsky.social. Do soutěže přihlásily snímky, které propojují krásu květu fuchsie se světem půdních bakterií Streptomyces coelicolor.



August 29, 2025 at 9:32 AM
Dvě vědkyně z @mbuavcr.bsky.social - Natalia Slonská a Lucie Najmanová, se zúčastnily soutěže Věda fotogenická, kterou pořádá @akademievedcr.bsky.social. Do soutěže přihlásily snímky, které propojují krásu květu fuchsie se světem půdních bakterií Streptomyces coelicolor.
Reposted by Michaela Novotná
Structures redefine the mechanism of action for tetracyclines https://www.biorxiv.org/content/10.1101/2025.09.25.678686v1
September 27, 2025 at 3:17 AM
Structures redefine the mechanism of action for tetracyclines https://www.biorxiv.org/content/10.1101/2025.09.25.678686v1
Reposted by Michaela Novotná
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
September 13, 2025 at 12:05 AM
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
Reposted by Michaela Novotná
Check out our new paper: a review of translational coupling, the phenomenon where translation of one prokaryotic gene can promote translation of the gene downstream. We cover the history, and delve into the mechanism, which is still not fully understood. journals.asm.org/doi/10.1128/...

Translational coupling of neighboring genes in prokaryotes | Journal of Bacteriology
Prokaryotic genes are arranged in operons, with functionally related genes often located
adjacent to one another (1). There are several ways in which the operonic organization of genes facilitates
the...
journals.asm.org
September 11, 2025 at 4:45 PM
Check out our new paper: a review of translational coupling, the phenomenon where translation of one prokaryotic gene can promote translation of the gene downstream. We cover the history, and delve into the mechanism, which is still not fully understood. journals.asm.org/doi/10.1128/...
Reposted by Michaela Novotná
Mechanism of 30S subunit recognition and modification by the conserved bacterial ribosomal RNA methyltransferase RsmI #microsky #rnasky 🦠 www.biorxiv.org/content/10.1...

Mechanism of 30S subunit recognition and modification by the conserved bacterial ribosomal RNA methyltransferase RsmI
Ribosomal RNA (rRNA) modifications are important for ribosome function and can influence bacterial susceptibility to ribosome-targeting antibiotics. The universally conserved 16S rRNA nucleotide C1402...
www.biorxiv.org
August 30, 2025 at 5:55 PM
Mechanism of 30S subunit recognition and modification by the conserved bacterial ribosomal RNA methyltransferase RsmI #microsky #rnasky 🦠 www.biorxiv.org/content/10.1...
Reposted by Michaela Novotná
Structural insights into YheS-mediated release of SecM-arrested ribosome. https://www.biorxiv.org/content/10.1101/2025.08.28.672766v1
August 31, 2025 at 5:46 AM
Structural insights into YheS-mediated release of SecM-arrested ribosome. https://www.biorxiv.org/content/10.1101/2025.08.28.672766v1
Reposted by Michaela Novotná
OMG this is it?! The final piece in the puzzle?!😲 tRNAs are the key, because archaic tRNAs sit at the core of all ribosomes. If aminoacyl-tRNAs could form on early Earth, you have a plausible route for the first self-replicators, the bridge from chemistry to biology
www.nature.com/articles/d41...
www.nature.com/articles/d41...

Origins of life: the molecules that could have unlocked peptide synthesis
For life to emerge on Earth, peptides must first have formed without the aid of enzymes — but how? Reactions of sulfur-containing molecules might have been key.
www.nature.com
August 28, 2025 at 8:00 AM
OMG this is it?! The final piece in the puzzle?!😲 tRNAs are the key, because archaic tRNAs sit at the core of all ribosomes. If aminoacyl-tRNAs could form on early Earth, you have a plausible route for the first self-replicators, the bridge from chemistry to biology
www.nature.com/articles/d41...
www.nature.com/articles/d41...
Reposted by Michaela Novotná
Our #CryoET story on how a top antimalarial drug candidate perturbs the native malarial translation machinery is out @natsmb.nature.com🥳 rdcu.be/eBbrH
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo

Integrated structural biology of the native malarial translation machinery and its inhibition by an antimalarial drug
Nature Structural & Molecular Biology - Integrated structural biology approach leveraging in situ cryo-electron tomography reveals molecular details of the native malarial translation...
rdcu.be
August 18, 2025 at 12:13 PM
Our #CryoET story on how a top antimalarial drug candidate perturbs the native malarial translation machinery is out @natsmb.nature.com🥳 rdcu.be/eBbrH
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
Happy to share a new paper from our lab with my little contribution! 🥳🥳🥳
We show that antibiotic resistance ABC-F proteins fine-tune responses to ribosome-binding antibiotics. Thanks to ABC-Fs, bacteria can differentiate between antibiotic classes and even concentrations for a tailored response. 🦠💊⬇️
We show that antibiotic resistance ABC-F proteins fine-tune responses to ribosome-binding antibiotics. Thanks to ABC-Fs, bacteria can differentiate between antibiotic classes and even concentrations for a tailored response. 🦠💊⬇️
New ABCF article from @balikovalab.bsky.social group!
Even though ABCF proteins have the same shape and act on the ribosome E-site, they have very different role in bacteria physiology level.
What an interesting topic!🤩
doi.org/10.1128/mbio...
Even though ABCF proteins have the same shape and act on the ribosome E-site, they have very different role in bacteria physiology level.
What an interesting topic!🤩
doi.org/10.1128/mbio...

ABCF protein-mediated resistance shapes bacterial responses to antibiotics based on their type and concentration | mBio
Bacteria adapt to diverse stimuli mainly through transcriptional changes that regulate adaptive protein factors. Here, we show that responses to protein synthesis-inhibiting antibiotics are fine-tuned by antibiotic resistance ABCF proteins at the ...
doi.org
August 13, 2025 at 11:54 AM
Happy to share a new paper from our lab with my little contribution! 🥳🥳🥳
We show that antibiotic resistance ABC-F proteins fine-tune responses to ribosome-binding antibiotics. Thanks to ABC-Fs, bacteria can differentiate between antibiotic classes and even concentrations for a tailored response. 🦠💊⬇️
We show that antibiotic resistance ABC-F proteins fine-tune responses to ribosome-binding antibiotics. Thanks to ABC-Fs, bacteria can differentiate between antibiotic classes and even concentrations for a tailored response. 🦠💊⬇️
Reposted by Michaela Novotná
Finally out!
I’m thrilled to share our new paper (Wolin et al., Cell 2025).
This paper describes SPIDR, a high-throughput method for mapping RBP binding sites.
By combining #SPIDR with #cryoEM, we identified the exact binding site of LARP1 within the #mRNA channel of the 40S ribosomal subunit.
I’m thrilled to share our new paper (Wolin et al., Cell 2025).
This paper describes SPIDR, a high-throughput method for mapping RBP binding sites.
By combining #SPIDR with #cryoEM, we identified the exact binding site of LARP1 within the #mRNA channel of the 40S ribosomal subunit.
July 22, 2025 at 5:12 PM
Reposted by Michaela Novotná
Out now:
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
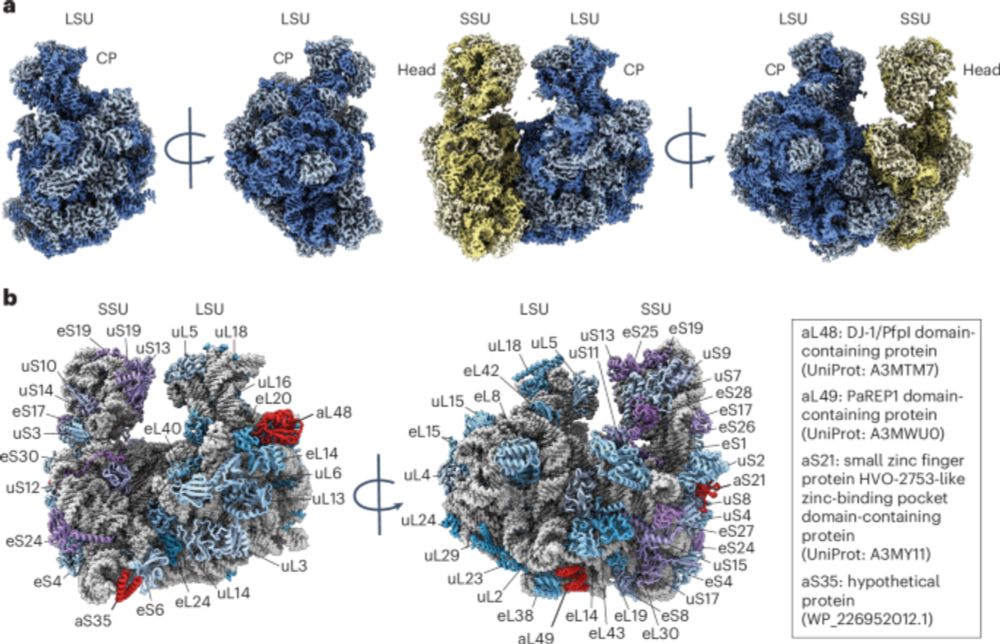
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor - Nature Microbiology
Sequence and structural analyses reveal a divergent peptidyl transferase centre and a hibernation factor in archaea.
www.nature.com
July 18, 2025 at 11:02 AM
Out now:
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor
@jhdcate.bsky.social @banfieldlab.bsky.social @diptinayak.bsky.social & co
#microsky
www.nature.com/articles/s41...
😲 E-site binding antibiotic discovered
👏👏
👏👏
Preprint from our collaboration with the labs of Gerry Wright and Shura Mankin identifying the first translation inhibitor binding to the E-site of the bacterial ribosome… www.researchsquare.com/article/rs-6...
A Natural Depsipeptide Antibiotic that Targets the E site of the Bacterial Ribosome
A significant challenge in addressing the antibiotic resistance crisis is identifying new antimicrobial compounds. Although natural products produced by fungi and bacteria, particularly actinomycetes,...
www.researchsquare.com
July 14, 2025 at 1:34 PM
😲 E-site binding antibiotic discovered
👏👏
👏👏
Reposted by Michaela Novotná
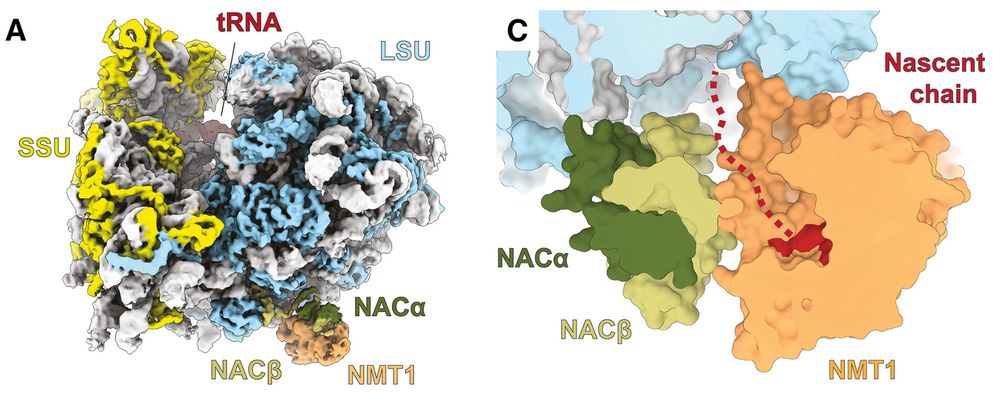
New paper out in Mol Cell, where we show how N-myristolytransferases lipidate proteins on the translating ribosome!
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...
July 12, 2025 at 12:10 AM
New paper out in Mol Cell, where we show how N-myristolytransferases lipidate proteins on the translating ribosome!
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...
Reposted by Michaela Novotná
Structural studies on ribosomes of differentially macrolide-resistant Staphylococcus aureus strains pubmed.ncbi.nlm.nih.gov/40490363/ #cryoem
June 10, 2025 at 3:48 PM
Structural studies on ribosomes of differentially macrolide-resistant Staphylococcus aureus strains pubmed.ncbi.nlm.nih.gov/40490363/ #cryoem
Reposted by Michaela Novotná
In situ protein crystallography: a single-crystal electron diffraction pipeline for structure determination inside living cells. www.biorxiv.org/content/10.1101/2025.07.08.663504v1 #cryoem
July 12, 2025 at 7:10 AM
In situ protein crystallography: a single-crystal electron diffraction pipeline for structure determination inside living cells. www.biorxiv.org/content/10.1101/2025.07.08.663504v1 #cryoem
Reposted by Michaela Novotná
Insights on how ribosomal RNA modification distant from drug binding site can still affect drug binding, in mycobacterium 🦠 #rnasky #ribosome #microsky
Distant ribose 2'-O-methylation of 23S rRNA helix 69 pre-orders the capreomycin drug binding pocket at the ribosome subunit interface pubmed.ncbi.nlm.nih.gov/40626557/ #cryoem
July 10, 2025 at 7:33 AM
Reposted by Michaela Novotná
Happy to say that my Current Opinion in Structural Biology article came out today. I hope it helps to guide to you in using cryo-ET to make new biological discoveries!
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...

A practical look at cryo-electron tomography image processing: Key considerations for new biological discoveries
Cryo-electron tomography (cryo-ET) enables 3D visualization of complex biological environments without the need for purification, thereby preserving t…
www.sciencedirect.com
July 8, 2025 at 9:44 PM
Happy to say that my Current Opinion in Structural Biology article came out today. I hope it helps to guide to you in using cryo-ET to make new biological discoveries!
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
Reposted by Michaela Novotná
Check out our recent review discussing the merits of tilt series vs single micrographs for in situ cryo-EM! www.sciencedirect.com/science/arti...

To tilt or not to tilt? Strategies for in situ cryo-EM data collection
Recent breakthroughs in single-particle cryogenic electron microscopy (cryo-EM) and protein structure prediction have transformed our ability to resol…
www.sciencedirect.com
June 28, 2025 at 2:04 PM
Check out our recent review discussing the merits of tilt series vs single micrographs for in situ cryo-EM! www.sciencedirect.com/science/arti...
Reposted by Michaela Novotná
Beautiful paper on translation start sites and extented SD motifs in Staph. aureus. www.biorxiv.org/content/10.1...
Congrats to colleagues in Strasburg univ.!
Congrats to colleagues in Strasburg univ.!

Extended Shine-Dalgarno motifs govern translation initiation in Staphylococcus aureus
Regulation of translation initiation is central to bacterial adaptation, but species-specific mechanisms remain poorly understood. We present high-resolution mapping of translation start sites in S. a...
www.biorxiv.org
June 27, 2025 at 1:05 PM
Beautiful paper on translation start sites and extented SD motifs in Staph. aureus. www.biorxiv.org/content/10.1...
Congrats to colleagues in Strasburg univ.!
Congrats to colleagues in Strasburg univ.!
Reposted by Michaela Novotná
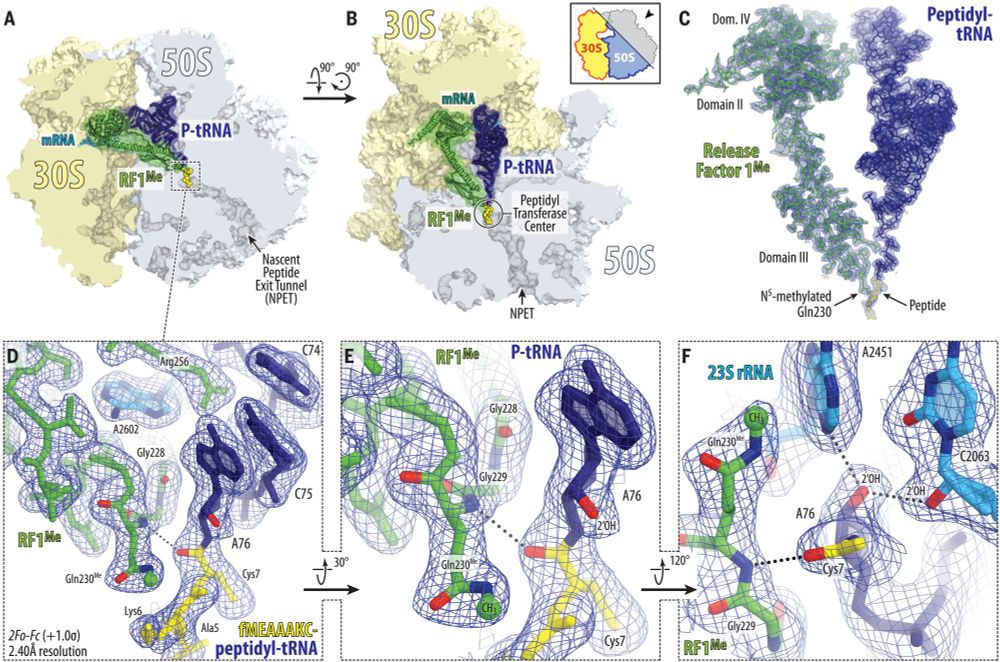
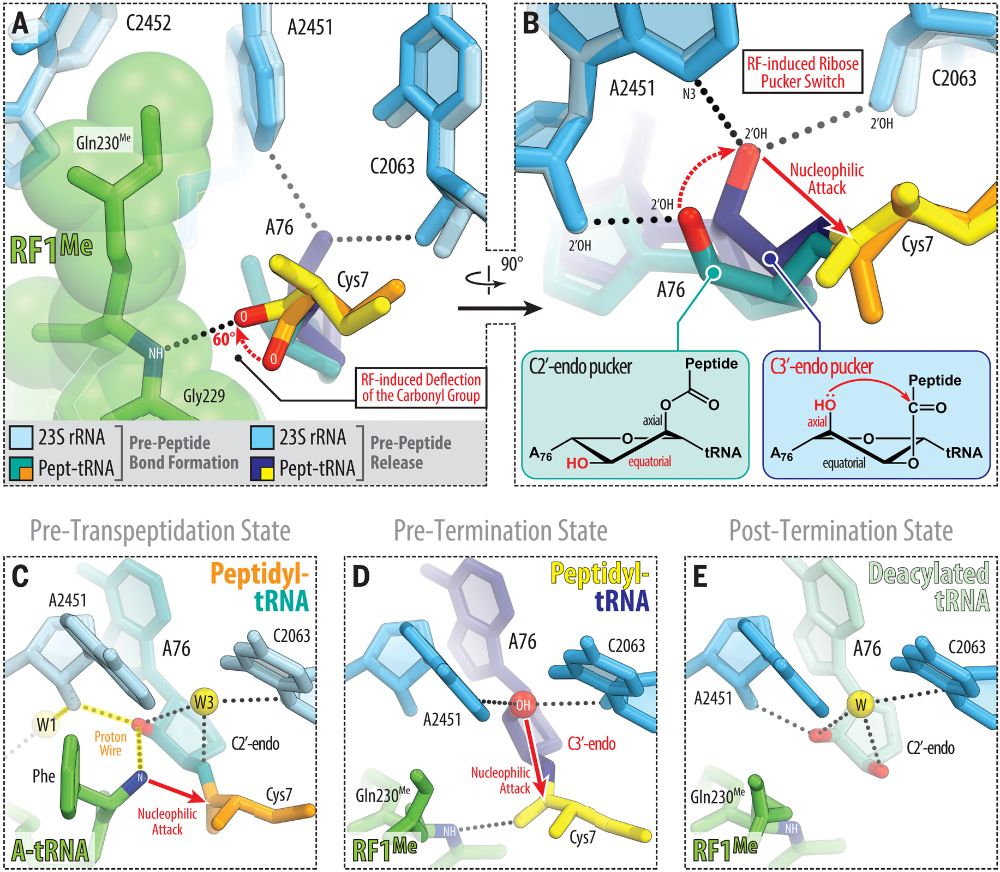
@science.org Mechanism of release factor–mediated peptidyl-tRNA hydrolysis on the ribosome | Science www.science.org/doi/10.1126/...
#ribosome #RNA #hydrolysis
#ribosome #RNA #hydrolysis
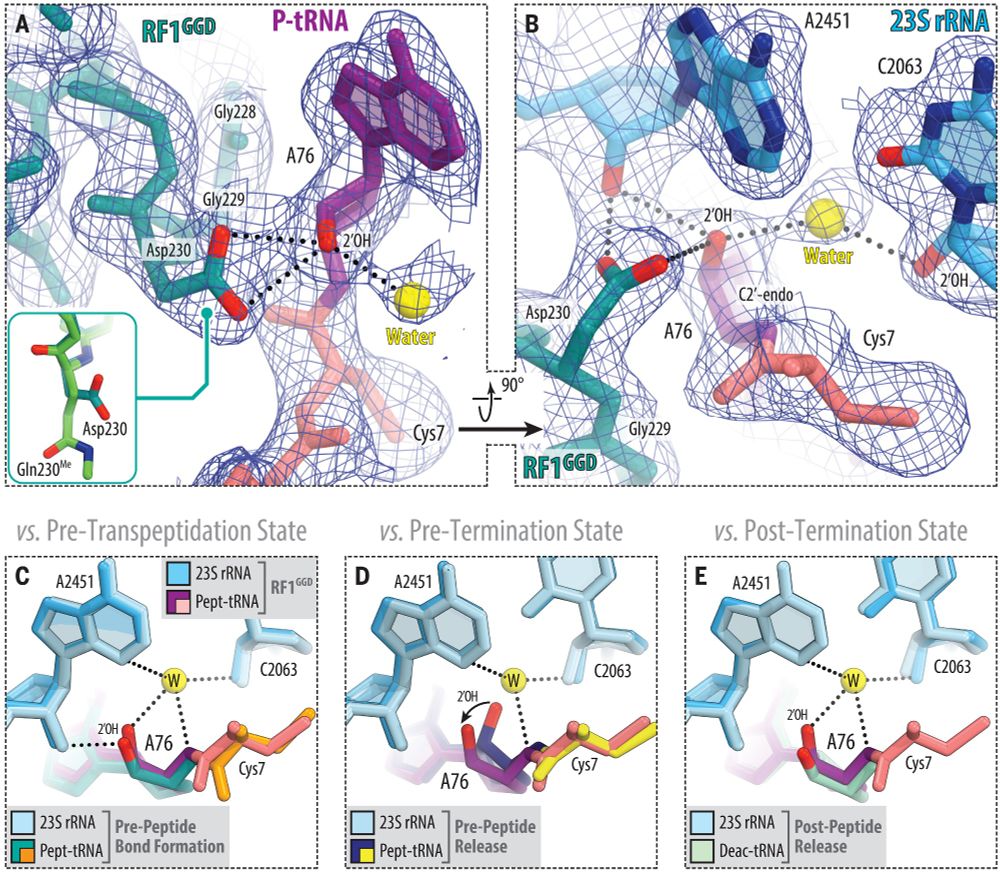
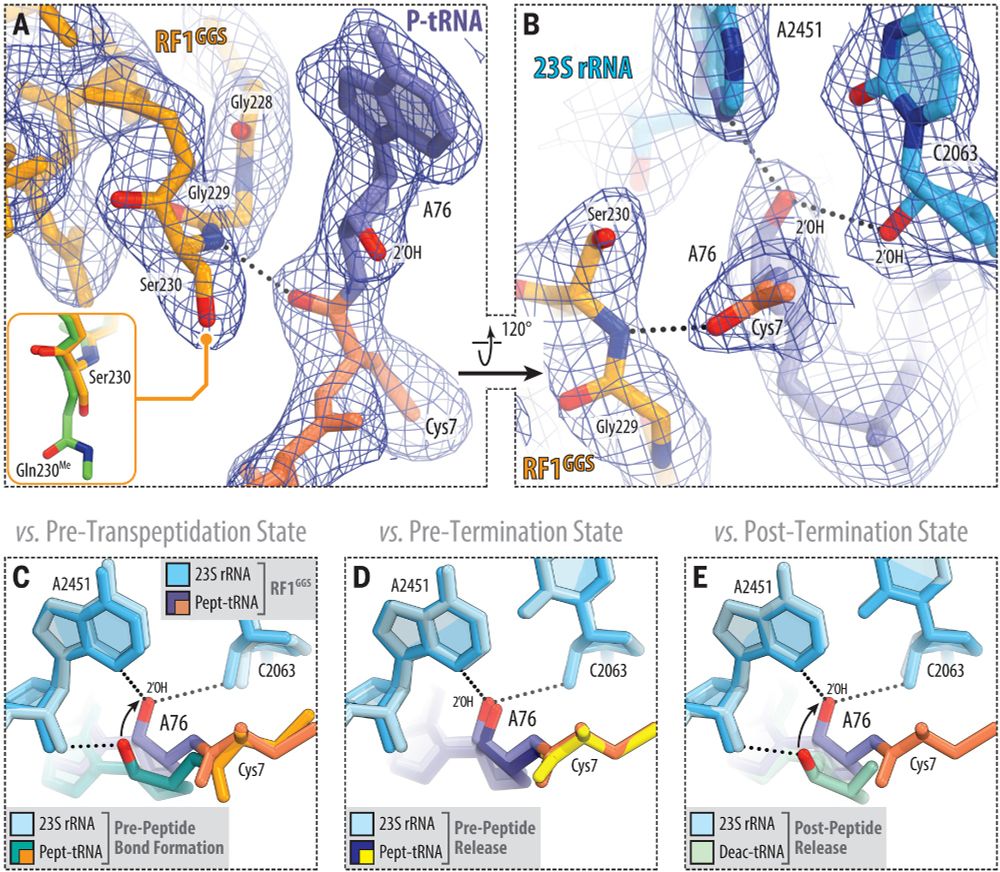
June 25, 2025 at 5:44 AM
@science.org Mechanism of release factor–mediated peptidyl-tRNA hydrolysis on the ribosome | Science www.science.org/doi/10.1126/...
#ribosome #RNA #hydrolysis
#ribosome #RNA #hydrolysis


