
computational biology | disease genetics
@mbadonyi.bsky.social @jmarshlab.bsky.social

📄 nature.com/articles/s41...



📄 onlinelibrary.wiley.com/doi/epdf/10....
📄 onlinelibrary.wiley.com/doi/epdf/10....
for the Mutational Scanning Symposium! #VariantEffect26
🗓️ March 25-27, 2026
📍 Melbourne, Australia (and online!)
📝 Submit your abstract (deadline is Nov 2nd, 2025!)
➡️ Registration is also now open!!
ℹ️ www.mss2026.org
#FunctionalGenomics #Symposium #Australia #CallForAbstracts

📄 nature.com/articles/s41...
@mbadonyi.bsky.social @jmarshlab.bsky.social

📄 nature.com/articles/s41...
This means long-term stability and easy installation with:
𝚒𝚗𝚜𝚝𝚊𝚕𝚕.𝚙𝚊𝚌𝚔𝚊𝚐𝚎𝚜('𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛')
🗞️ doi.org/10.1093/bioi...
#rstats #acmg #varianteffect #MAVEs #VEPs #genomics

This means long-term stability and easy installation with:
𝚒𝚗𝚜𝚝𝚊𝚕𝚕.𝚙𝚊𝚌𝚔𝚊𝚐𝚎𝚜('𝚊𝚌𝚖𝚐𝚜𝚌𝚊𝚕𝚎𝚛')
🗞️ doi.org/10.1093/bioi...
#rstats #acmg #varianteffect #MAVEs #VEPs #genomics
The R package now has a single calibrate() function, and the Colab interface is easier to use.
📄 Manuscript: www.biorxiv.org/content/10.1...
🧪 Colab: edin.ac/4mjzijp
#rstats @theacmg.bsky.social

The R package now has a single calibrate() function, and the Colab interface is easier to use.
📄 Manuscript: www.biorxiv.org/content/10.1...
🧪 Colab: edin.ac/4mjzijp
#rstats @theacmg.bsky.social
www.biorxiv.org/content/10.1...
Many thanks to all involved - @wbickmor.bsky.social @mbadonyi.bsky.social @eliasfriman.bsky.social, Joe Marsh, Jasmine Nguyen, Mark Gorrell and others.

www.biorxiv.org/content/10.1...
Many thanks to all involved - @wbickmor.bsky.social @mbadonyi.bsky.social @eliasfriman.bsky.social, Joe Marsh, Jasmine Nguyen, Mark Gorrell and others.
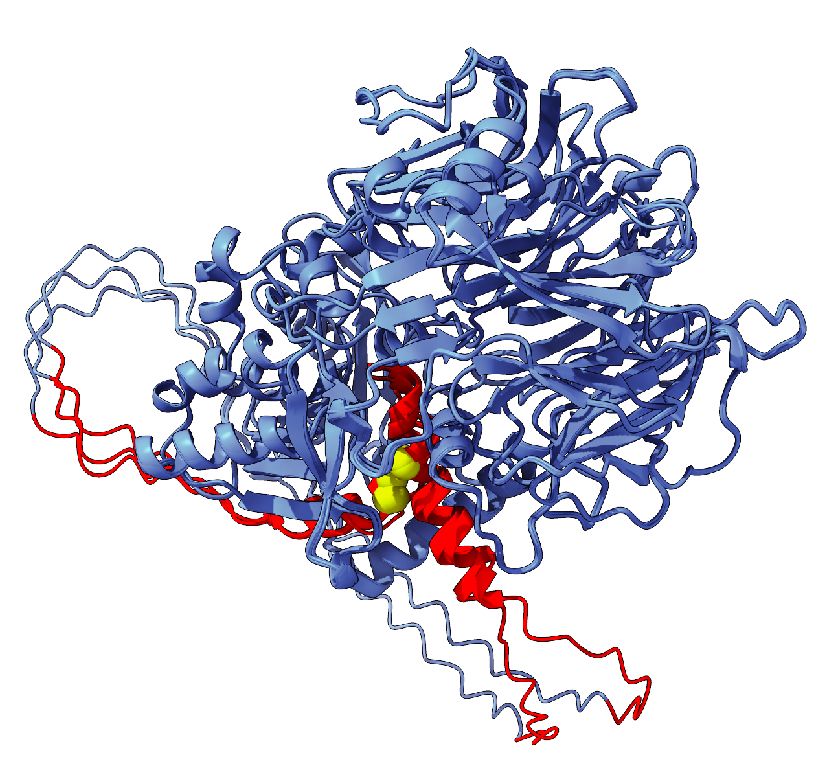
Your estimate is probably too high:

Your estimate is probably too high:
www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics

www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics
We lay out best-practice guidelines for releasing variant effect predictors, developed through the Atlas of Variant Effects Alliance @varianteffect.bsky.social
Open, interpretable, and clinically useful VEPs are the goal.
📄 doi.org/10.1186/s130...

We took a photo of her and her little collaborator! Her key words are #BabiesInScience #PUFFFIN #NeighbourLabelling. Check out this interview with Tamina: thenode.biologists.com/the-sdb-bsdb...

We took a photo of her and her little collaborator! Her key words are #BabiesInScience #PUFFFIN #NeighbourLabelling. Check out this interview with Tamina: thenode.biologists.com/the-sdb-bsdb...


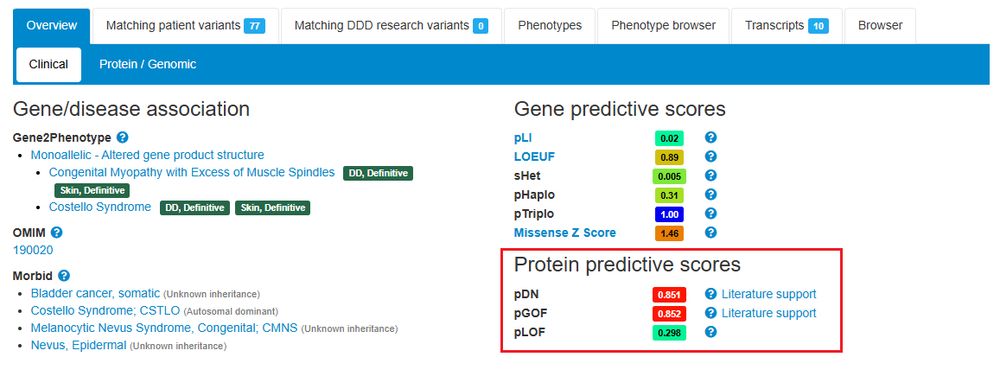

I have also created an R Colab for these solutions, so R-curious people can run them without having to install R: github.com/badonyi/adve...

I have also created an R Colab for these solutions, so R-curious people can run them without having to install R: github.com/badonyi/adve...




