



Have a question you'd like me to answer during my session? Drop it in the comments or message me, I'd love to hear your thoughts.
Further information: hubs.la/Q03JMvwd0
#FOGLondon #genomics #biodata

Have a question you'd like me to answer during my session? Drop it in the comments or message me, I'd love to hear your thoughts.
Further information: hubs.la/Q03JMvwd0
#FOGLondon #genomics #biodata
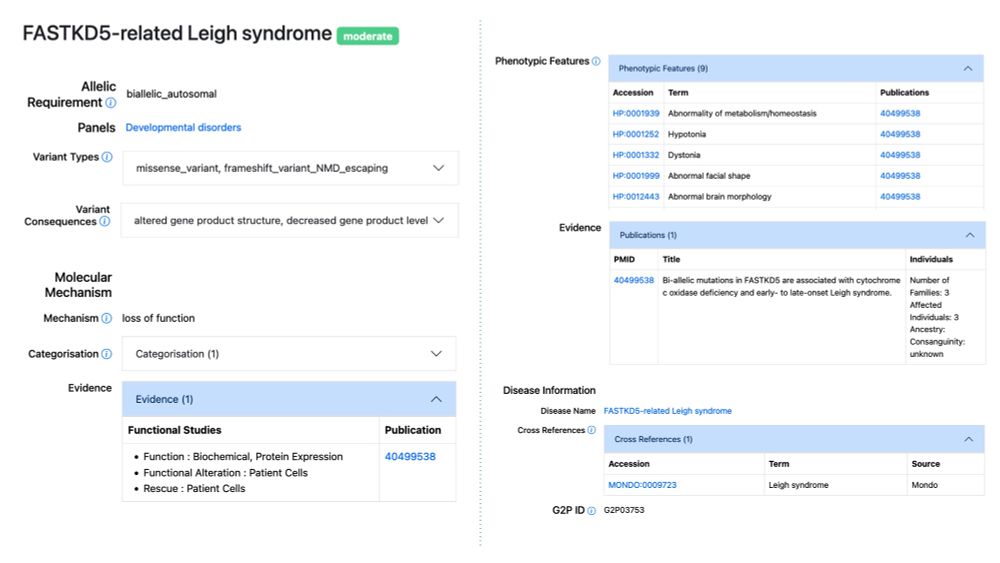
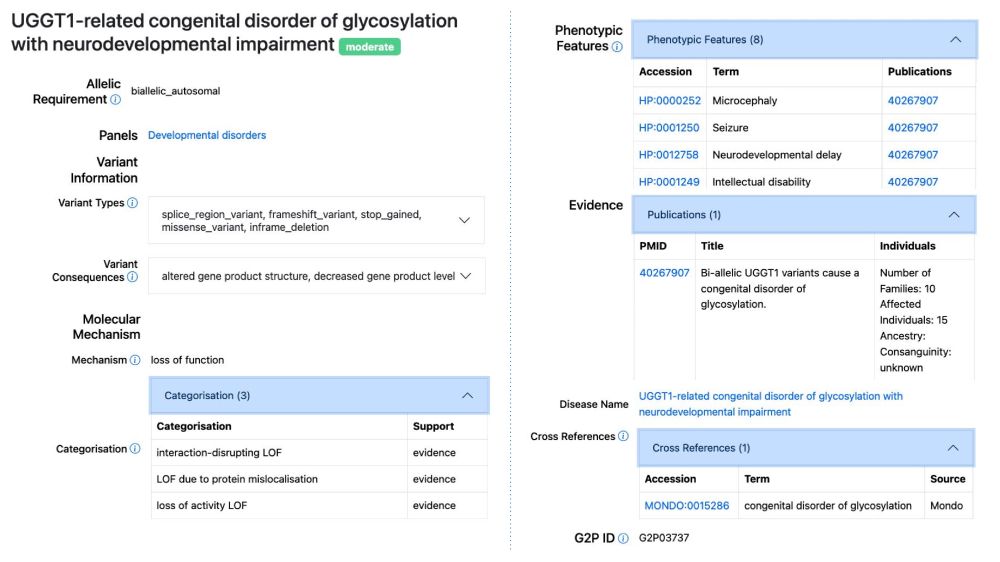
Publications identified using a new machine learning approach can now be searched and browsed in G2P.
www.ebi.ac.uk/about/news/u...

Publications identified using a new machine learning approach can now be searched and browsed in G2P.
www.ebi.ac.uk/about/news/u...



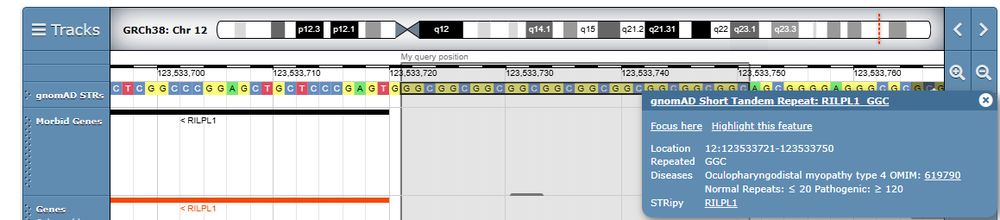
A new method, LMSz, creates condition-specific growth charts and is being integrated in @deciphergenomics.bsky.social
www.ebi.ac.uk/about/news/t...
🧬💻

A new method, LMSz, creates condition-specific growth charts and is being integrated in @deciphergenomics.bsky.social
www.ebi.ac.uk/about/news/t...
🧬💻







🗓️ Starts: 7 July
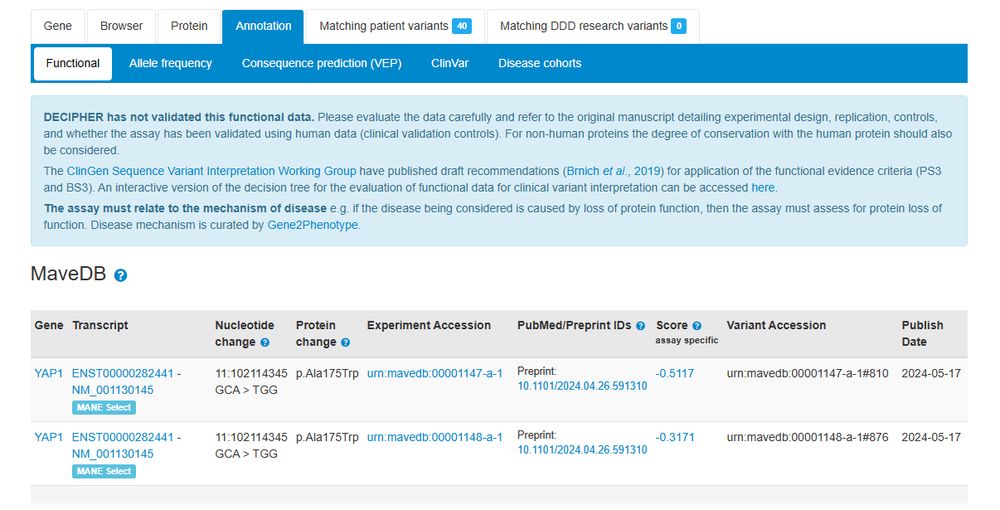
Learn how to get the most from #genomics data from diverse populations, using variant classification and interpretation approaches. #GenomeVariation🧬
📎bit.ly/3R7G7aK

🗓️ Starts: 7 July
Learn how to get the most from #genomics data from diverse populations, using variant classification and interpretation approaches. #GenomeVariation🧬
📎bit.ly/3R7G7aK

