
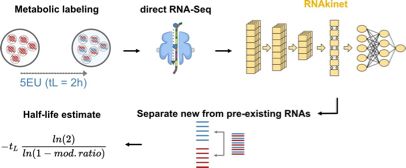
#RNAsky

#RNAsky
@kevinrhine.bsky.social
www.nature.com/articles/s41...
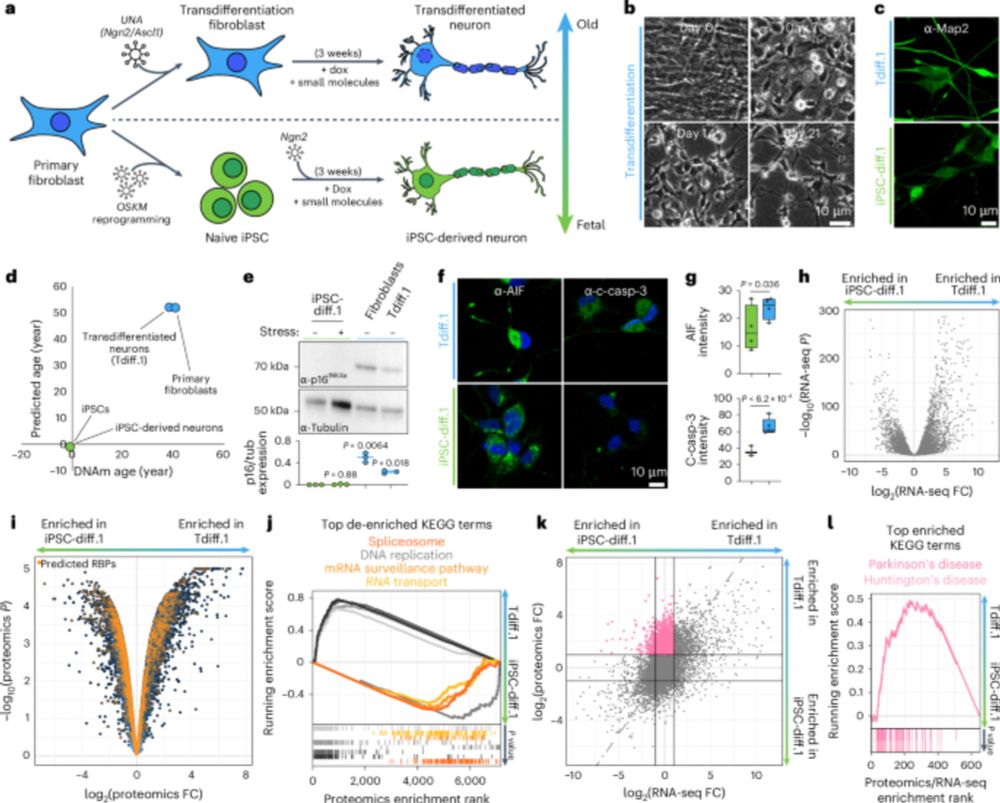
@kevinrhine.bsky.social
www.nature.com/articles/s41...
Passionate about #RNA biology, #MachineLearning, and the #genomics of #aging?
Interested in a postdoc or a PhD research visit at NIH? Let’s connect.
📩 DM or email for details!
#Bioinformatics #AI #ComputationalBiology
Passionate about #RNA biology, #MachineLearning, and the #genomics of #aging?
Interested in a postdoc or a PhD research visit at NIH? Let’s connect.
📩 DM or email for details!
#Bioinformatics #AI #ComputationalBiology
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
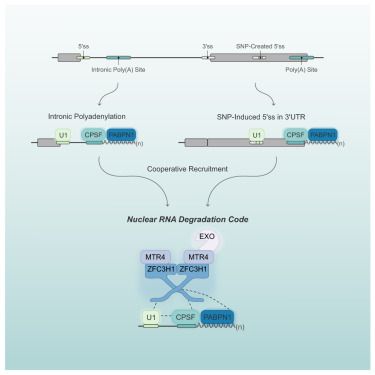
#RNAsky
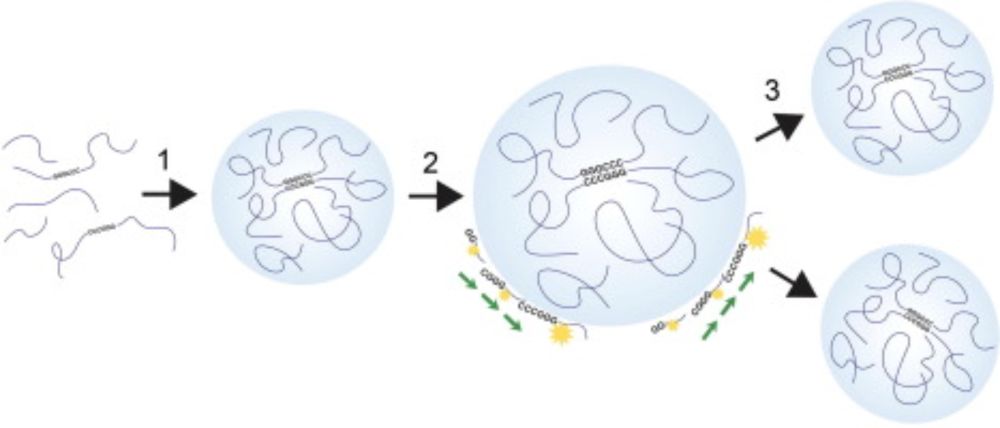
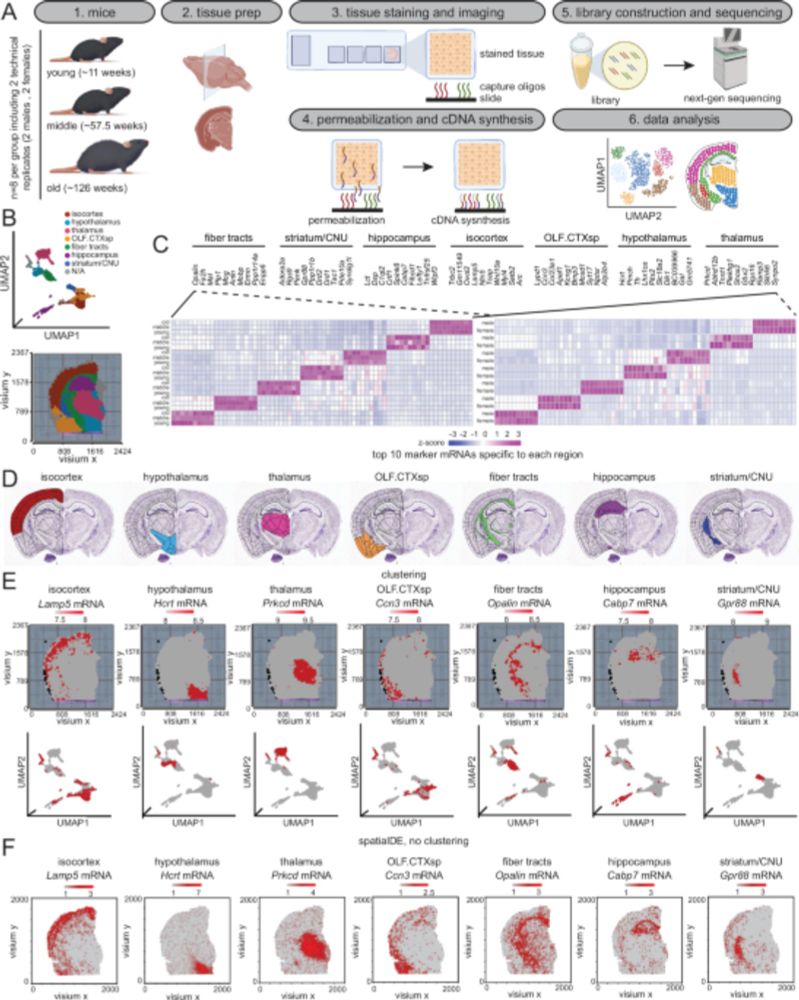
Great work from @ferhatalkan.bsky.social, Edwin Kyei-Baffour, and Qi Chang Lin!
royalsocietypublishing.org/doi/10.1098/...
Great work from @ferhatalkan.bsky.social, Edwin Kyei-Baffour, and Qi Chang Lin!
royalsocietypublishing.org/doi/10.1098/...


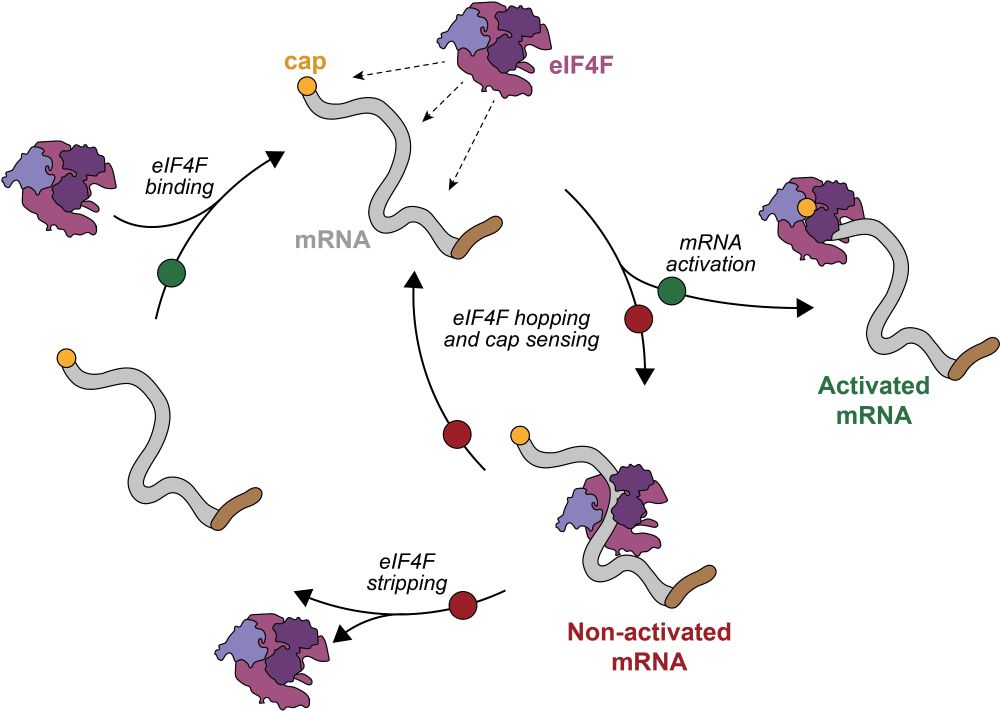
🤔 useful to further reduce translational load during #ISR?
#RNAbiology

🤔 useful to further reduce translational load during #ISR?
#RNAbiology


We normally don't ask you to "share like and subscribe" but please distribute this.
You are now able to delete the link after it embeds on your post so you can get those precious extra characters, and we will catch your post mentioning research.
#AcademicSky #Medsky #HigherEd
We normally don't ask you to "share like and subscribe" but please distribute this.
You are now able to delete the link after it embeds on your post so you can get those precious extra characters, and we will catch your post mentioning research.
#AcademicSky #Medsky #HigherEd
#genomics #bioinformatics
#genomics #bioinformatics

Check the app here 👉 pagoba.shinyapps.io/strain_explo...
#AcademicSky
#Science

Check the app here 👉 pagoba.shinyapps.io/strain_explo...
#AcademicSky
#Science
#RNAsky
www.cell.com/molecular-ce...

#RNAsky
www.cell.com/molecular-ce...
www.nature.com/articles/s43...

www.nature.com/articles/s43...
doi.org/10.1038/s415...

doi.org/10.1038/s415...
In our new study, we ask the question: Can we predict RNA stability across conditions and protocols, based solely on experimental and/or computationally predicted RNA binding protein target sites?
Read the paper to find out !
www.biorxiv.org/content/10.1...

www.nature.com/articles/s41...
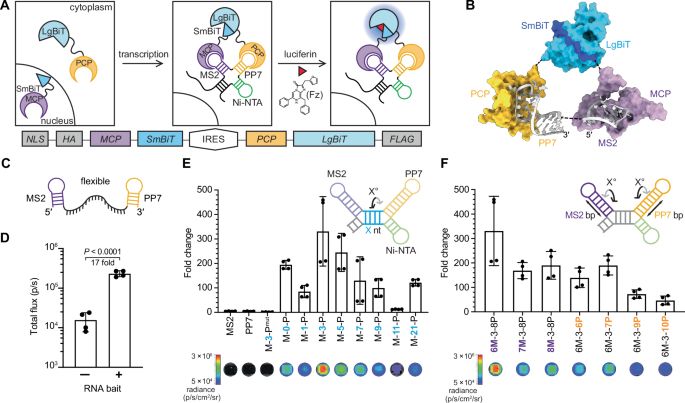
www.nature.com/articles/s41...

