We show why you should do genotype-level QC on your WGS data
www.biorxiv.org/content/10.1...
Very real quotes about this paper -
“The most exciting, mind-blowing paper of the year!”
“On a par with Fisher 1918”
“I read it every night. Just so beautiful”

We show why you should do genotype-level QC on your WGS data
www.biorxiv.org/content/10.1...
Very real quotes about this paper -
“The most exciting, mind-blowing paper of the year!”
“On a par with Fisher 1918”
“I read it every night. Just so beautiful”
A big step for precision medicine!
www.nature.com/articles/s41...

A big step for precision medicine!
www.nature.com/articles/s41...
(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...

(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...
Are you a bioinformatician / computational scientist who wants to apply your skills to understanding regulatory biology and improving rare disease diagnosis and treatment? 🧠 💻 🧬 🩺
We have two roles available 👇
🧵 1/4

Are you a bioinformatician / computational scientist who wants to apply your skills to understanding regulatory biology and improving rare disease diagnosis and treatment? 🧠 💻 🧬 🩺
We have two roles available 👇
🧵 1/4
Within only 145 nucleotides(!) of a non-coding RNA (RNU4-2) - different variants in distinct regions / structures cause three distinct disorders!!! (all discovered within the last 18 months)
🤯🤓🧬❤️

Within only 145 nucleotides(!) of a non-coding RNA (RNU4-2) - different variants in distinct regions / structures cause three distinct disorders!!! (all discovered within the last 18 months)
🤯🤓🧬❤️
"Who licks bone !?!" 🦴
- Johannes Krause
Anyone have that on your bingo card?
Well apparently archeologists do, to distinguish bone from stones and it causes problems in DNA sequencing. 🤔

"Who licks bone !?!" 🦴
- Johannes Krause
Anyone have that on your bingo card?
Well apparently archeologists do, to distinguish bone from stones and it causes problems in DNA sequencing. 🤔
I know multiple of these have already occurred.
Paying a lot of attention to the entire event will pay off!

I know multiple of these have already occurred.
Packed program of excellent science 8.30am-8.00pm - plus networking event till 9.30pm to meet many friends, colleagues and collaborators! …andiamo @eshg.bsky.social @eshgyoung.bsky.social
Packed program of excellent science 8.30am-8.00pm - plus networking event till 9.30pm to meet many friends, colleagues and collaborators! …andiamo @eshg.bsky.social @eshgyoung.bsky.social
github.com/populationge...
A superb collaboration with @dgmacarthur.bsky.social @cassimons.bsky.social @heidirehm.bsky.social @ksamocha.bsky.social and many more!

github.com/populationge...
A superb collaboration with @dgmacarthur.bsky.social @cassimons.bsky.social @heidirehm.bsky.social @ksamocha.bsky.social and many more!

📅 Dates: 9-11 April 2025
💭 Share insights in person
Explore the latest #genomics advances accelerating improvements in clinical care for rare disorders, globally.
⏰Secure your place by 11 March: bit.ly/3BpAe44

📅 Dates: 9-11 April 2025
💭 Share insights in person
Explore the latest #genomics advances accelerating improvements in clinical care for rare disorders, globally.
⏰Secure your place by 11 March: bit.ly/3BpAe44
www.biorxiv.org/content/10.1...
1/10
www.biorxiv.org/content/10.1...
1/10
The @gnomad-project.bsky.social team is hard at work on v4.1 and improvements across the board, so expect more updates.
Thanks to Katherine Chao for spearheading this blogpost.
The @gnomad-project.bsky.social team is hard at work on v4.1 and improvements across the board, so expect more updates.
Thanks to Katherine Chao for spearheading this blogpost.
We hope that the resource we generated will be useful when interpreting rare co-occurring variants in the context of recessive disease.
www.nature.com/articles/s41...
We hope that the resource we generated will be useful when interpreting rare co-occurring variants in the context of recessive disease.
www.nature.com/articles/s41...
1/6
1/6
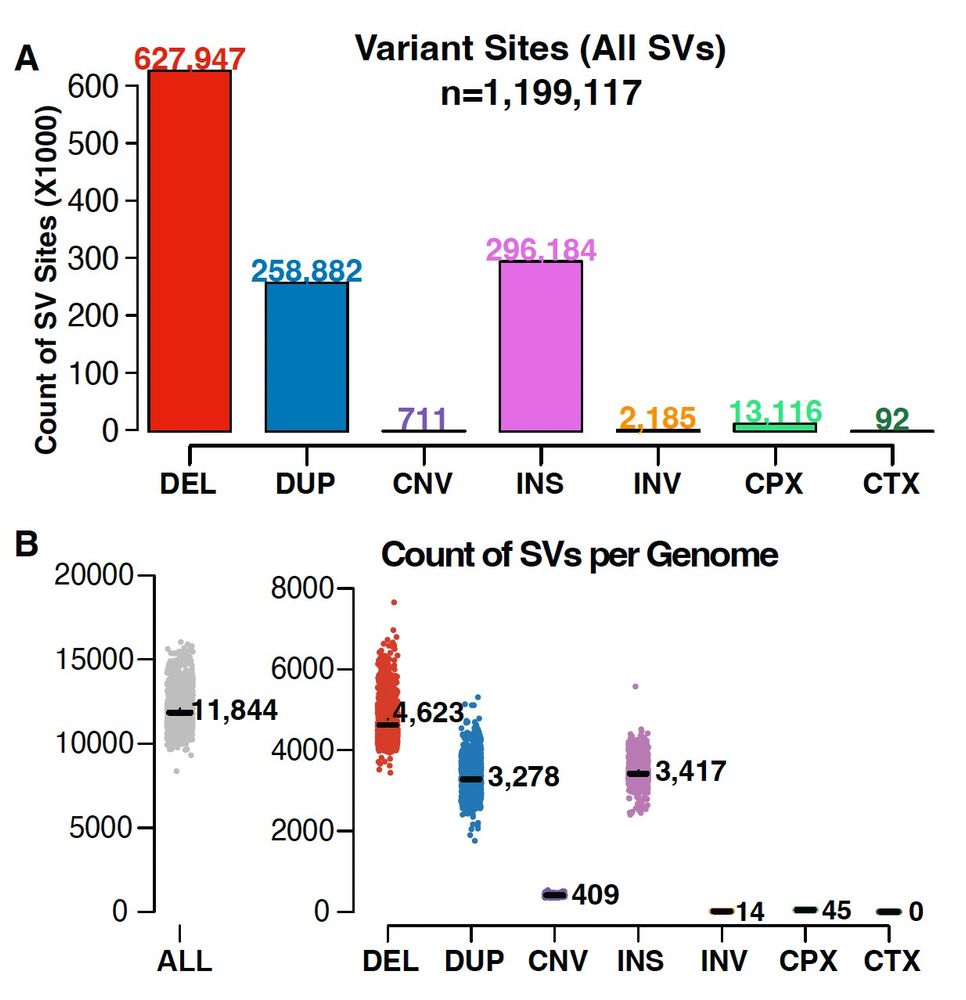
v4 is the first time we released structural variants at the same time as SNVs/indels, specifically:
- CNVs from 464,297 exomes
- SVs from 63,046 genomes
1/4

v4 is the first time we released structural variants at the same time as SNVs/indels, specifically:
- CNVs from 464,297 exomes
- SVs from 63,046 genomes
1/4
As a reminder, I'm only highlighting a few individuals of the many that contribute. You can see more on our team page:
gnomad.broadinstitute.org/team
1/5

As a reminder, I'm only highlighting a few individuals of the many that contribute. You can see more on our team page:
gnomad.broadinstitute.org/team
1/5
With 150k+ views a week, the browser is a crucial part of making gnomAD accessible. Quickly loading data from >800k samples + presenting it in a user-friendly format is no small feat.
1/6

With 150k+ views a week, the browser is a crucial part of making gnomAD accessible. Quickly loading data from >800k samples + presenting it in a user-friendly format is no small feat.
1/6
First up this week is the amazing production team.
1/7

First up this week is the amazing production team.
1/7