I view this iteration as more of a franchise reboot instead of a sequel – we have new leads, but similar themes.
9/10


I view this iteration as more of a franchise reboot instead of a sequel – we have new leads, but similar themes.
9/10
8/10
8/10
7/10
7/10
6/10
6/10
5/10

5/10
4/10
4/10
3/10
3/10
2/10
2/10
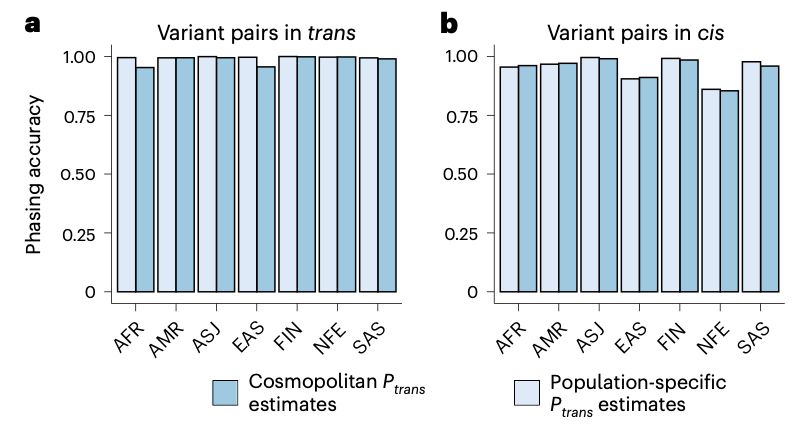
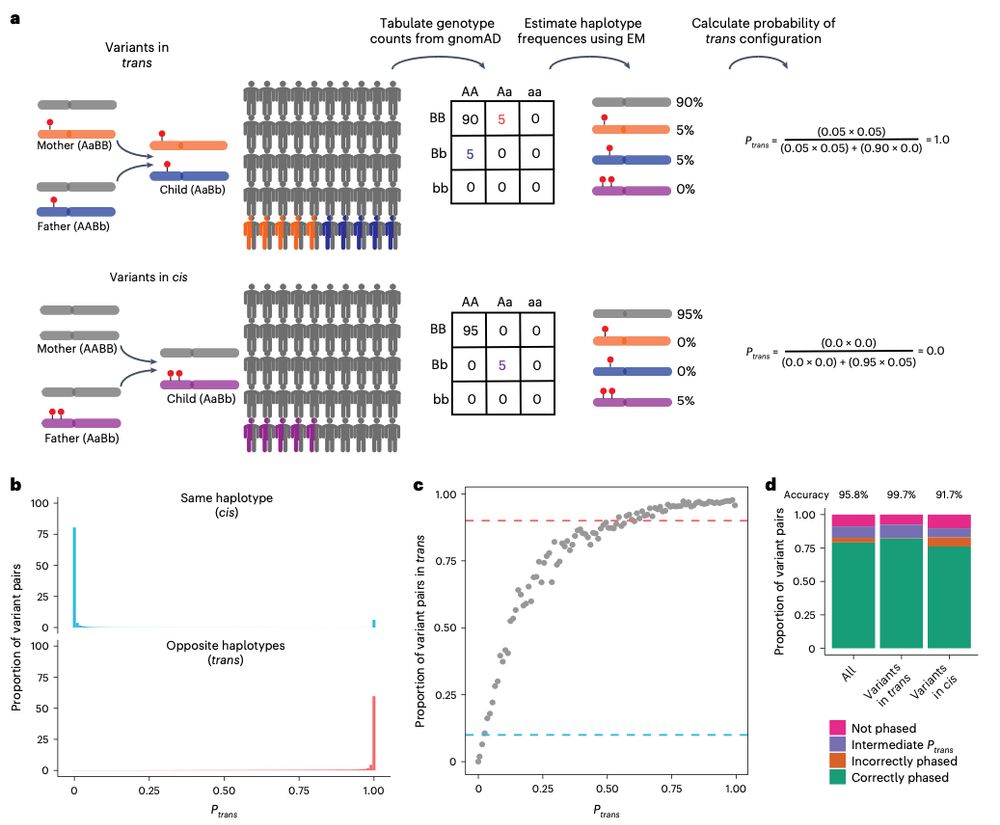
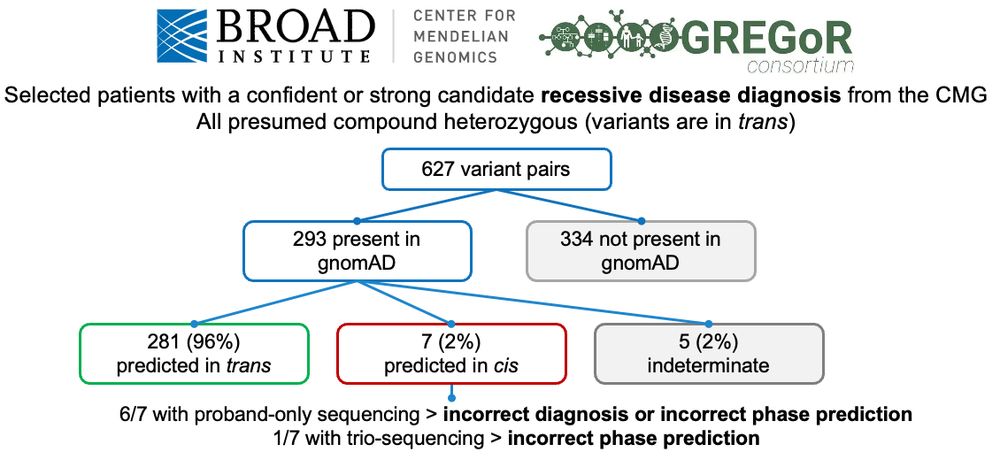
5/6

5/6
4/6

4/6
To all of you: thank you for contributing. You are the heart of gnomAD.
2/6

To all of you: thank you for contributing. You are the heart of gnomAD.
2/6
v4 is the first time we released structural variants at the same time as SNVs/indels, specifically:
- CNVs from 464,297 exomes
- SVs from 63,046 genomes
1/4

v4 is the first time we released structural variants at the same time as SNVs/indels, specifically:
- CNVs from 464,297 exomes
- SVs from 63,046 genomes
1/4
As a reminder, I'm only highlighting a few individuals of the many that contribute. You can see more on our team page:
gnomad.broadinstitute.org/team
1/5

As a reminder, I'm only highlighting a few individuals of the many that contribute. You can see more on our team page:
gnomad.broadinstitute.org/team
1/5
6/6

6/6
5/6

5/6
gnomad.broadinstitute.org/news/2023-11...
He also recently added the variant co-occurrence counts table to the gene pages in v2.
4/6

gnomad.broadinstitute.org/news/2023-11...
He also recently added the variant co-occurrence counts table to the gene pages in v2.
4/6
With 150k+ views a week, the browser is a crucial part of making gnomAD accessible. Quickly loading data from >800k samples + presenting it in a user-friendly format is no small feat.
1/6

With 150k+ views a week, the browser is a crucial part of making gnomAD accessible. Quickly loading data from >800k samples + presenting it in a user-friendly format is no small feat.
1/6
First up this week is the amazing production team.
1/7

First up this week is the amazing production team.
1/7


