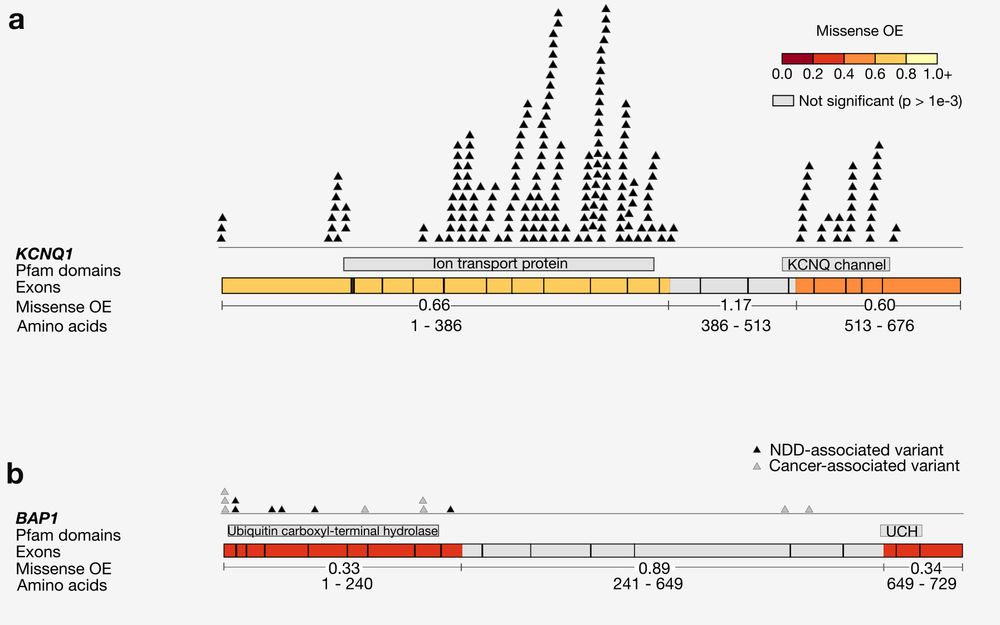
Not sure if recommended, but it saved time.
Not sure if recommended, but it saved time.
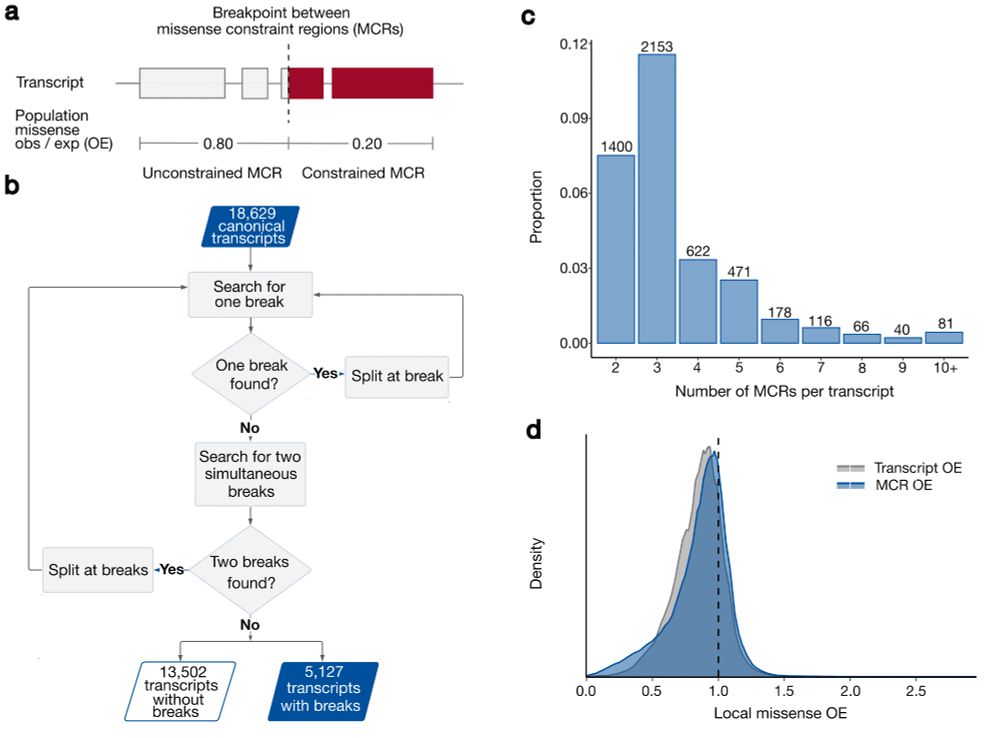
10/10
10/10
I view this iteration as more of a franchise reboot instead of a sequel – we have new leads, but similar themes.
9/10


I view this iteration as more of a franchise reboot instead of a sequel – we have new leads, but similar themes.
9/10
8/10
8/10
7/10
7/10
6/10
6/10
5/10

5/10
4/10
4/10
3/10
3/10
2/10
2/10

