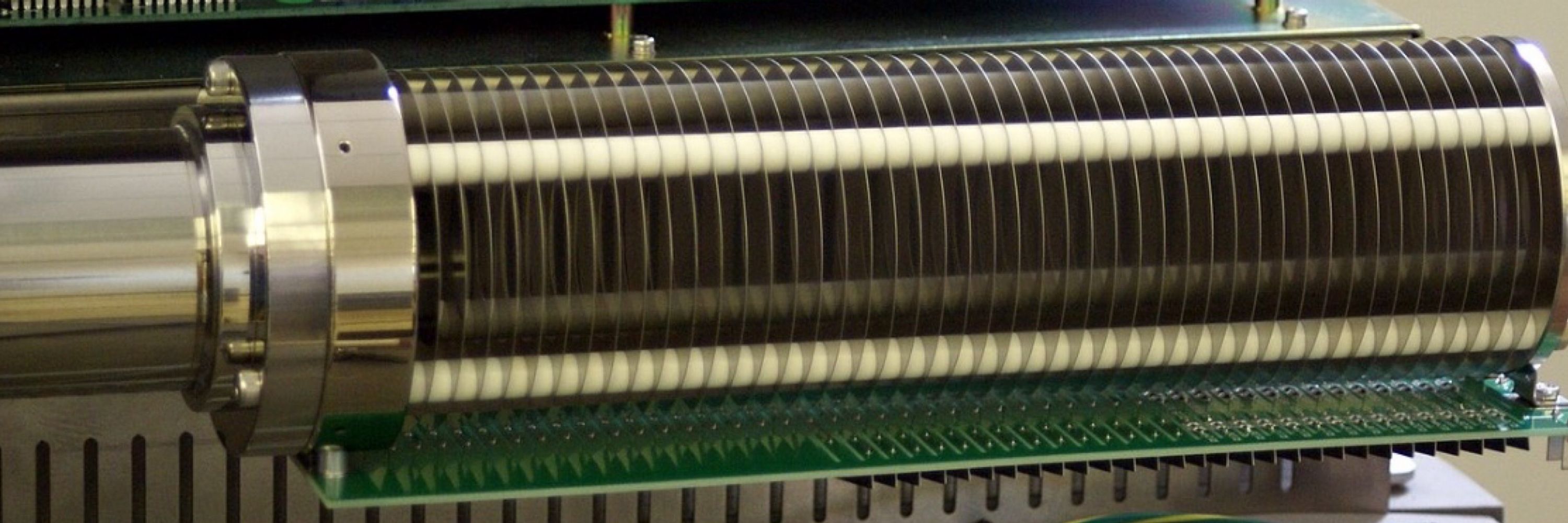
“We have developed an electron-activated dissociation (EAD) device with product isolation functionality for multistage tandem mass spectrometry (MSn)”
Hopefully commercially available soon, the ability to do MSn is invaluable in characterisation workflows.
“We have developed an electron-activated dissociation (EAD) device with product isolation functionality for multistage tandem mass spectrometry (MSn)”
Hopefully commercially available soon, the ability to do MSn is invaluable in characterisation workflows.

It's called EU Tenders (ted.europa.eu) and has many recent MS prices on there.
E.g. Astral Zoom with FAIMS Pro Duo is listed as $1.75 million Euro ($2 million USD)
Limited info but probably includes an unlisted LC
It's called EU Tenders (ted.europa.eu) and has many recent MS prices on there.
E.g. Astral Zoom with FAIMS Pro Duo is listed as $1.75 million Euro ($2 million USD)
Limited info but probably includes an unlisted LC
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Let’s see if Waters can make a big dent in the proteomics field with this

Let’s see if Waters can make a big dent in the proteomics field with this


Our open-source DIA framework brings deep learning directly to raw MS data with feature-free processing, transfer learning for any PTM, and performance matching top tools.
www.nature.com/articles/s41...

Our open-source DIA framework brings deep learning directly to raw MS data with feature-free processing, transfer learning for any PTM, and performance matching top tools.
www.nature.com/articles/s41...

It automates the manual work of extracting method parameters, presenting them in a structured and comparable view.
(A thread 🧵)

It automates the manual work of extracting method parameters, presenting them in a structured and comparable view.
(A thread 🧵)
If we can get vendor buy-in to an open, common format, it could bring more computational folks into the field. How can these folks be excited about #proteomics when its hard to even read the already complicated data?
pubs.acs.org/doi/10.1021/...

Analyses macromolecules from 100 MDa and beyond. No charge state deconvolution necessary.
www.waters.com/nextgen/au/e...

Analyses macromolecules from 100 MDa and beyond. No charge state deconvolution necessary.
www.waters.com/nextgen/au/e...

One sphere is black (1 ppm).
Finding the black sphere is comparable to detecting a protein present at ~ 6,000 copies in the proteome of a human cell.
Quantifying the protein requires analyzing multiple jars.

One sphere is black (1 ppm).
Finding the black sphere is comparable to detecting a protein present at ~ 6,000 copies in the proteome of a human cell.
Quantifying the protein requires analyzing multiple jars.
This could also work for the Orbitrap Astral.
Bonus: DIAPASEF on Thermo - patentscope.wipo.int/search/en/de...

This could also work for the Orbitrap Astral.
Bonus: DIAPASEF on Thermo - patentscope.wipo.int/search/en/de...
go.nature.com/4gHDSGF

go.nature.com/4gHDSGF
They should make their money back in other ways (software subscriptions in particular), but if you want the cheapest, single cell-level MS, the 8600 is probably it.

They should make their money back in other ways (software subscriptions in particular), but if you want the cheapest, single cell-level MS, the 8600 is probably it.
Nova - lightweight dev library
schweppelab.github.io/Nova/ pubs.acs.org/doi/full/10....
Helios - a unified instrument API now @ JPR
github.com/SchweppeLab/...

Nova - lightweight dev library
schweppelab.github.io/Nova/ pubs.acs.org/doi/full/10....
Helios - a unified instrument API now @ JPR
github.com/SchweppeLab/...
The excerpt is from: "Orbitrap Against All Odds"
theanalyticalscientist.com/issues/2013/...

The excerpt is from: "Orbitrap Against All Odds"
theanalyticalscientist.com/issues/2013/...

---
#proteomics #prot-paper

---
#proteomics #prot-paper
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
go.nature.com/40GxAjN

go.nature.com/40GxAjN

pubs.acs.org/doi/10.1021/...
This was the solution I found to be most useful.
pubs.acs.org/doi/10.1021/...
This was the solution I found to be most useful.

