India -> Australia
github.com/pwilmart/qua...
github.com/pwilmart/qua...
phbuffers.org/Claude/pepti...
phbuffers.org/Claude/pepti...




---
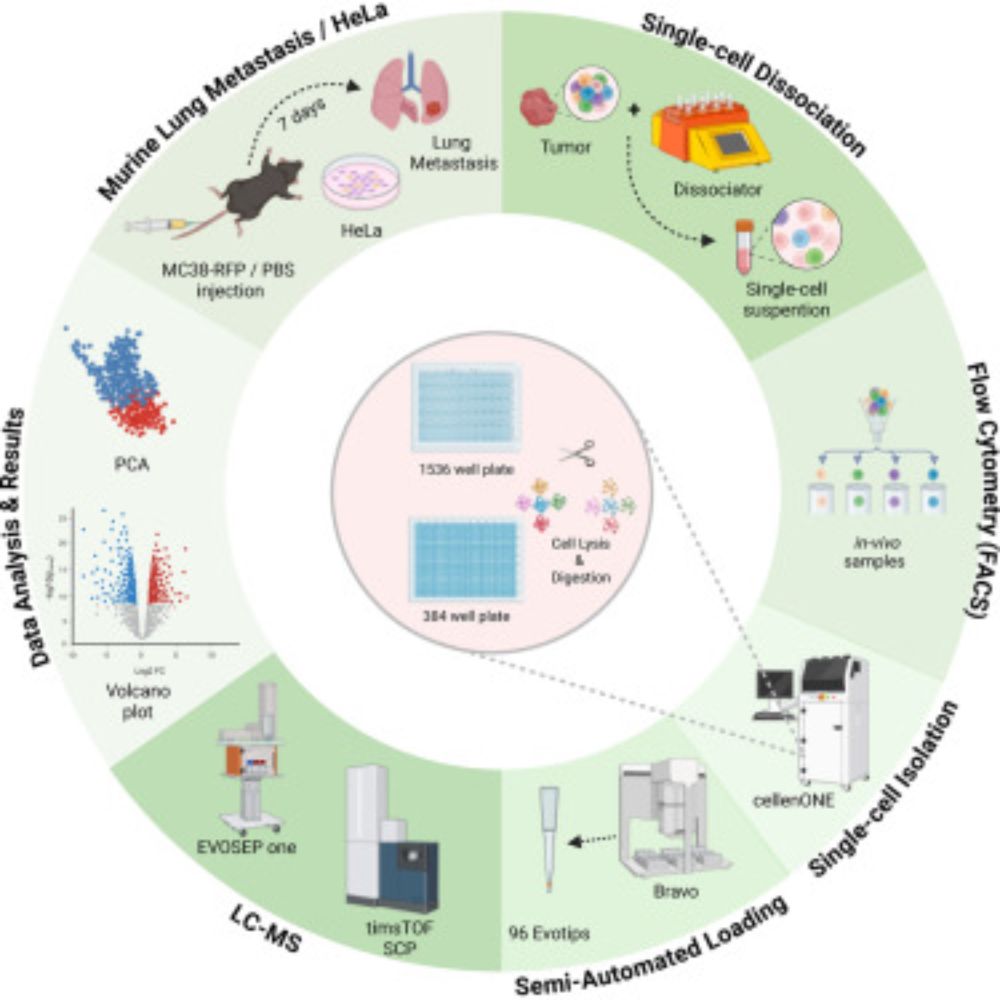
#proteomics #prot-preprint
---
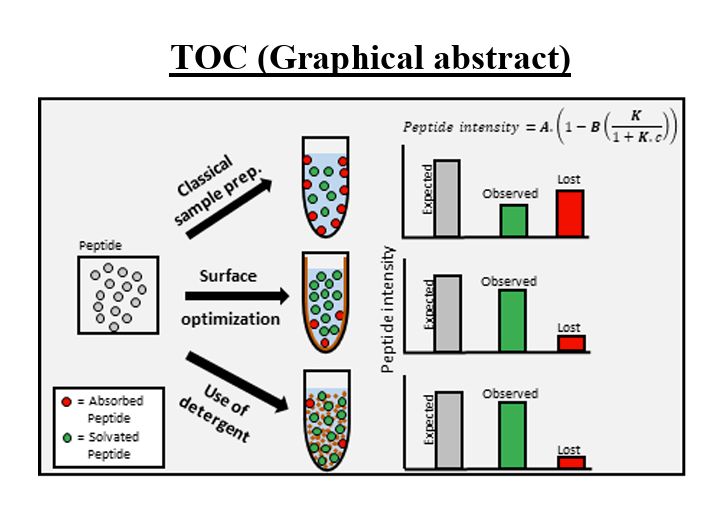
#proteomics #prot-preprint

---
#proteomics #prot-paper

---
#proteomics #prot-paper
www.cell.com/iscience/ful...
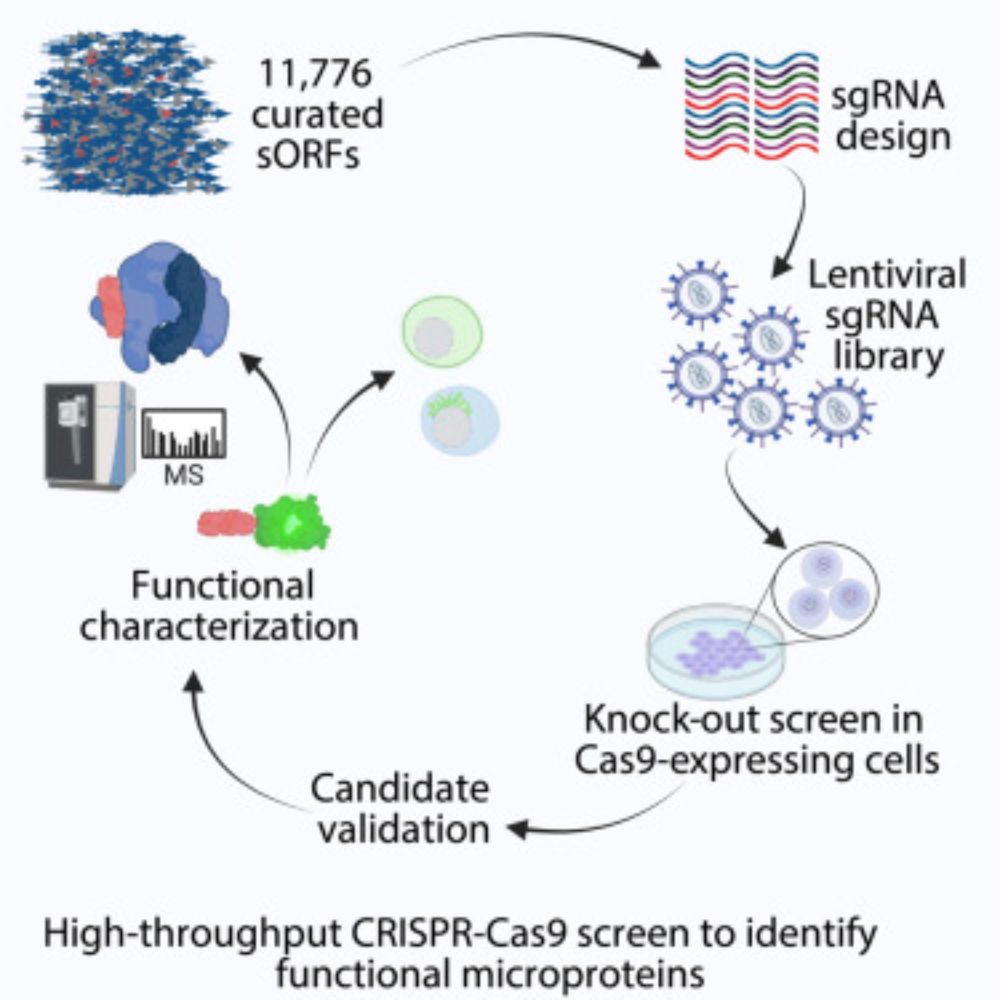
www.cell.com/iscience/ful...

---
#proteomics #prot-preprint
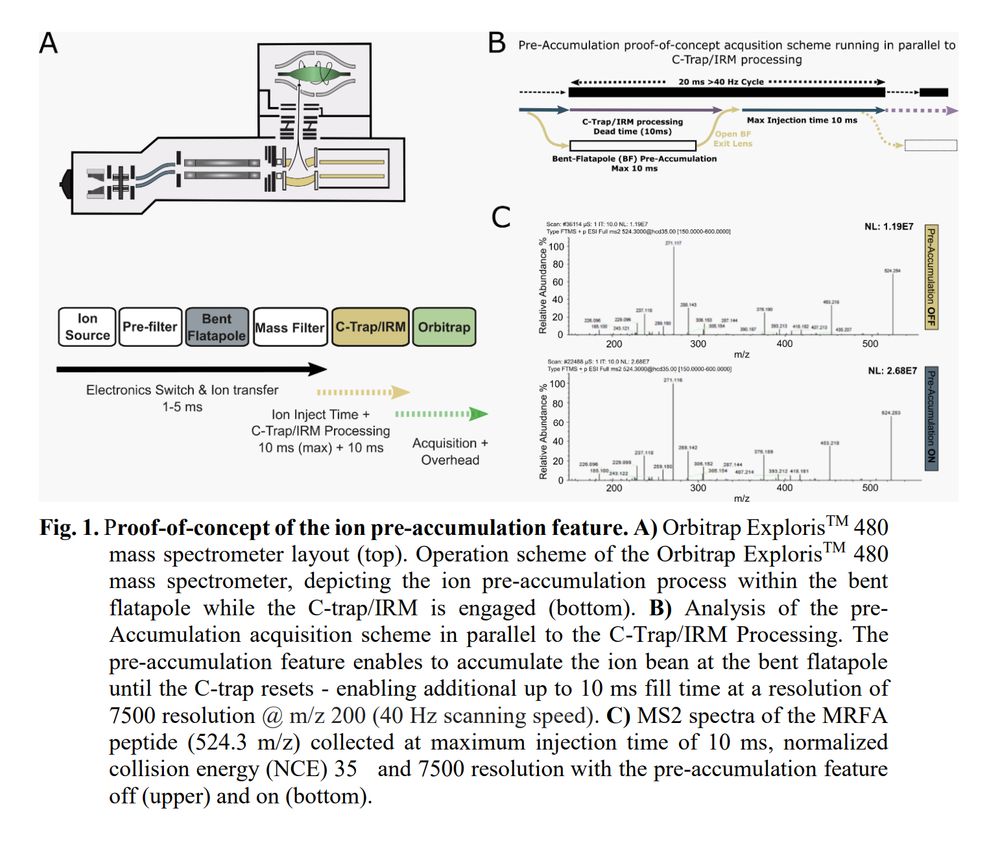
---
#proteomics #prot-preprint
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
www.jic.ac.uk/vacancies/pr...
Closing date - 2 March 2025
Contract - Full-time, indefinite

www.jic.ac.uk/vacancies/pr...
Closing date - 2 March 2025
Contract - Full-time, indefinite

Identification of O-glycopeptides from tandem mass spectrometry data is complicated by the near complete dissociation of O-glycans from the peptide during collisional… http://dlvr.it/THhR43 #MassSpecRSS

Identification of O-glycopeptides from tandem mass spectrometry data is complicated by the near complete dissociation of O-glycans from the peptide during collisional… http://dlvr.it/THhR43 #MassSpecRSS

Digitizing the human proteome in >53,000 individuals with 15-year follow-up and, with machine learning, unraveling nearly 500 disease-causing proteins
www.cell.com/cell/fulltex...
open-access

Digitizing the human proteome in >53,000 individuals with 15-year follow-up and, with machine learning, unraveling nearly 500 disease-causing proteins
www.cell.com/cell/fulltex...
open-access


