Chromatin is very dynamic and flexible, but NOT regular!!! 🧪🧬🔬
My career and work: http://bit.ly/2CuF4L5
Lab HP: https://bit.ly/3F1a8nk
YouTube Seminar: https://bit.ly/4fYZiOj
www.biorxiv.org/cgi/content/...
doi.org/10.1101/2025...
#Microscopy #CellBiology #OI-DIC #SMLM

doi.org/10.1101/2025...
#Microscopy #CellBiology #OI-DIC #SMLM
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
Also consistent with many previous studies.
(4/n)
Also consistent with many previous studies.
(4/n)
With @shiori-iida.bsky.social , We revealed the detailed structure of euchromatin using super-resolution imaging.
Cohesin regulates the function of euchromatin via chromatin dynamics!
(1/n)
www.biorxiv.org/cgi/content/...
With @shiori-iida.bsky.social , We revealed the detailed structure of euchromatin using super-resolution imaging.
Cohesin regulates the function of euchromatin via chromatin dynamics!
(1/n)
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
We combined single-nucleosome imaging and 3D-SIM to reveal:
🔹 Euchromatin forms condensed domains, not open fibers
🔹Cohesin loss increases nucleosome mobility without decompaction
🔹Cohesin prevents neighboring domain mixing
Full story & movies👇
www.biorxiv.org/cgi/content/...
We combined single-nucleosome imaging and 3D-SIM to reveal:
🔹 Euchromatin forms condensed domains, not open fibers
🔹Cohesin loss increases nucleosome mobility without decompaction
🔹Cohesin prevents neighboring domain mixing
Full story & movies👇
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...
Our study, utilizing embryonic samples from cooperative aquariums, revealed the ancient sex chromosome origin and unique sex determination potentials.
Published today in PNAS
www.pnas.org/doi/full/10....
#Squalomix
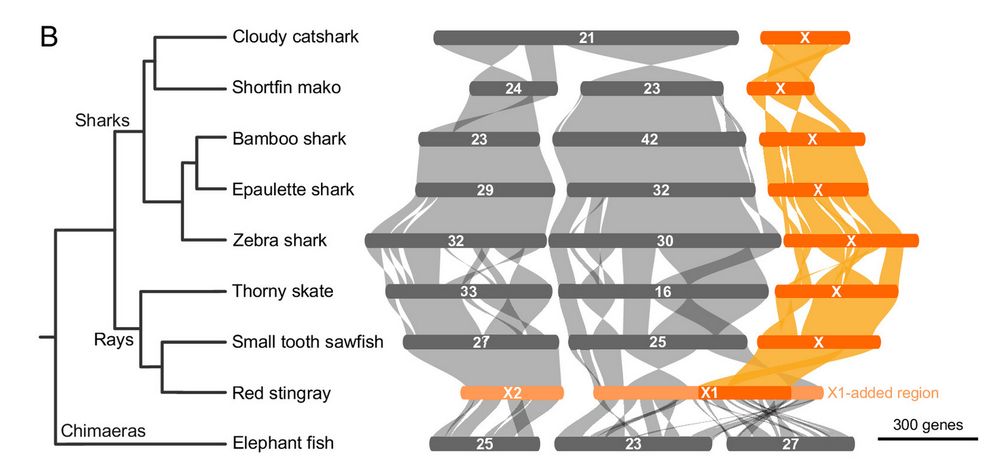
Our study, utilizing embryonic samples from cooperative aquariums, revealed the ancient sex chromosome origin and unique sex determination potentials.
Published today in PNAS
www.pnas.org/doi/full/10....
#Squalomix
Link: biorxiv.org/content/10.110…
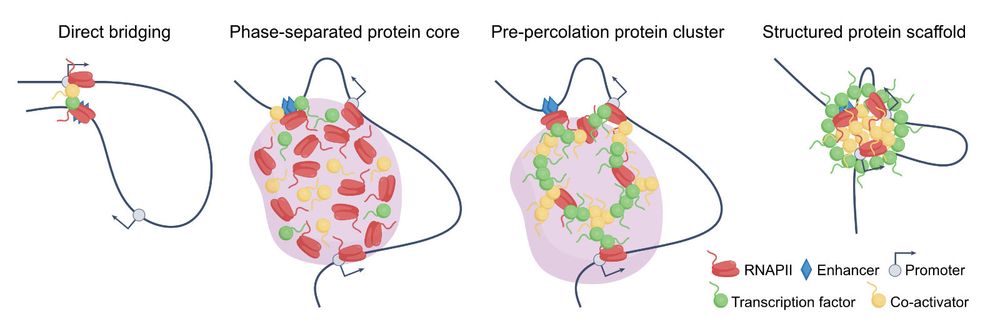
Using single-molecul#imagingng, we showed that liquid-like BRD4-NUT condensates locally constrai#chromatinin vi#bromodomain-dependentnt crosslinking.
I thank my supervisor, Prof. @kazu-maeshima.bsky.social, for his support!

Link: biorxiv.org/content/10.110…
Using single-molecul#imagingng, we showed that liquid-like BRD4-NUT condensates locally constrai#chromatinin vi#bromodomain-dependentnt crosslinking.
I thank my supervisor, Prof. @kazu-maeshima.bsky.social, for his support!
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
Excited to share our work on "Ab-trapping," an antibody artifact causing misleading peripheral ("rim") staining in imaging & genomics (IF, CUT&Tag, CUT&RUN). Antibodies fail to penetrate structures, accumulating at the periphery. A 🧵👇
doi.org/10.1101/2025...

Excited to share our work on "Ab-trapping," an antibody artifact causing misleading peripheral ("rim") staining in imaging & genomics (IF, CUT&Tag, CUT&RUN). Antibodies fail to penetrate structures, accumulating at the periphery. A 🧵👇
doi.org/10.1101/2025...
A short thread follows 1/n

A short thread follows 1/n
"Evidence suggests that acetyl-CoA metabolism is highly compartmentalized in mammalian cells. Yet methods to measure acetyl-CoA in living cells are lacking. Herein, we engineered an acetyl-CoA biosensor from the bacterial protein PanZ and circularly permuted green fluorescent protein (cpGFP)."

"Evidence suggests that acetyl-CoA metabolism is highly compartmentalized in mammalian cells. Yet methods to measure acetyl-CoA in living cells are lacking. Herein, we engineered an acetyl-CoA biosensor from the bacterial protein PanZ and circularly permuted green fluorescent protein (cpGFP)."
"Replication-dependent histone labeling dissects the physical properties of euchromatin/heterochromatin in living human cells"
From @kazu-maeshima.bsky.social lab
www.science.org/doi/10.1126/...

"Replication-dependent histone labeling dissects the physical properties of euchromatin/heterochromatin in living human cells"
From @kazu-maeshima.bsky.social lab
www.science.org/doi/10.1126/...
@katsuminami.bsky.social @semeigazin.bsky.social @kakonakazato.bsky.social discuss DNA accessibility in euchromatin and heterochromatin of live cells.
Free access: authors.elsevier.com/a/1lHgb54HFZ...
Also check our previous work: www.science.org/doi/10.1126/...
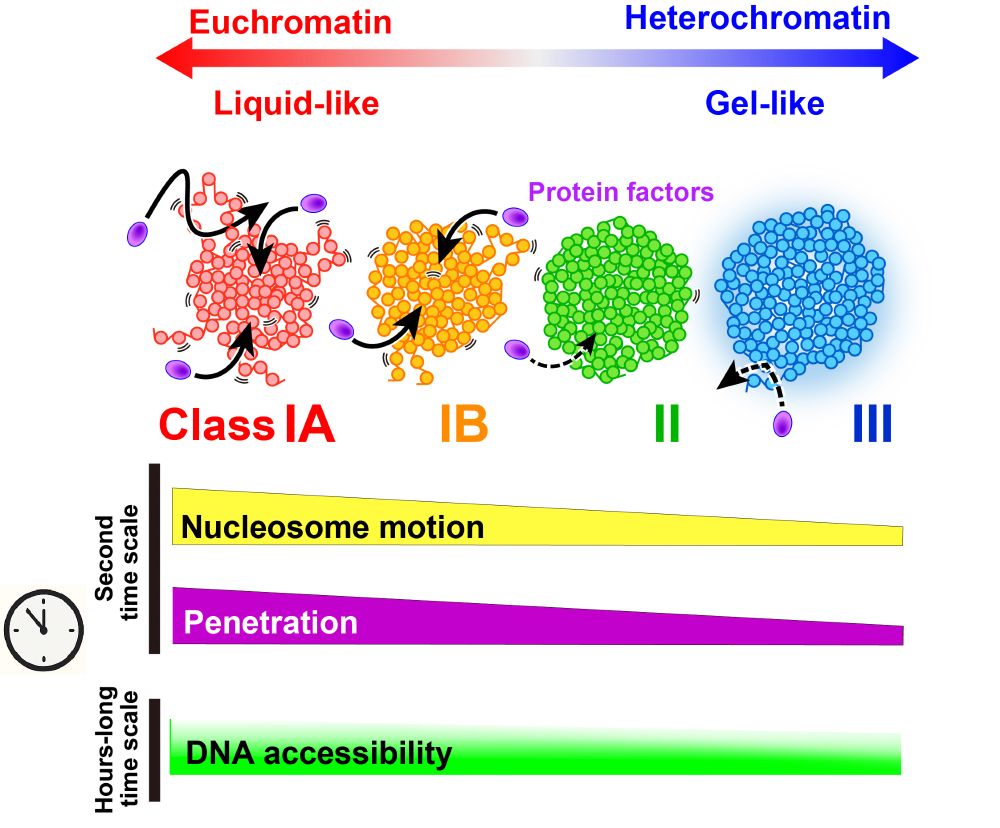
@katsuminami.bsky.social @semeigazin.bsky.social @kakonakazato.bsky.social discuss DNA accessibility in euchromatin and heterochromatin of live cells.
Free access: authors.elsevier.com/a/1lHgb54HFZ...
Also check our previous work: www.science.org/doi/10.1126/...
https://go.nature.com/3Zh7bs7

https://go.nature.com/3Zh7bs7
First ever studies of full length BRD4 with intact nucleosomes find that it binds with nanomolar affinity even in the ABSENCE of acetylation.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

First ever studies of full length BRD4 with intact nucleosomes find that it binds with nanomolar affinity even in the ABSENCE of acetylation.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
New preprint!
How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites?
www.biorxiv.org/content/10.1...
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics

New preprint!
How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites?
www.biorxiv.org/content/10.1...
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics
🎫 Register by 3 Jun 👉🏼 http://s.embl.org/grg25-01-bl
🧬RNA processing
🧬Translation
🧬Transcription & chromatin regulation
🧬Method development
🧬Theory

🎫 Register by 3 Jun 👉🏼 http://s.embl.org/grg25-01-bl
🧬RNA processing
🧬Translation
🧬Transcription & chromatin regulation
🧬Method development
🧬Theory
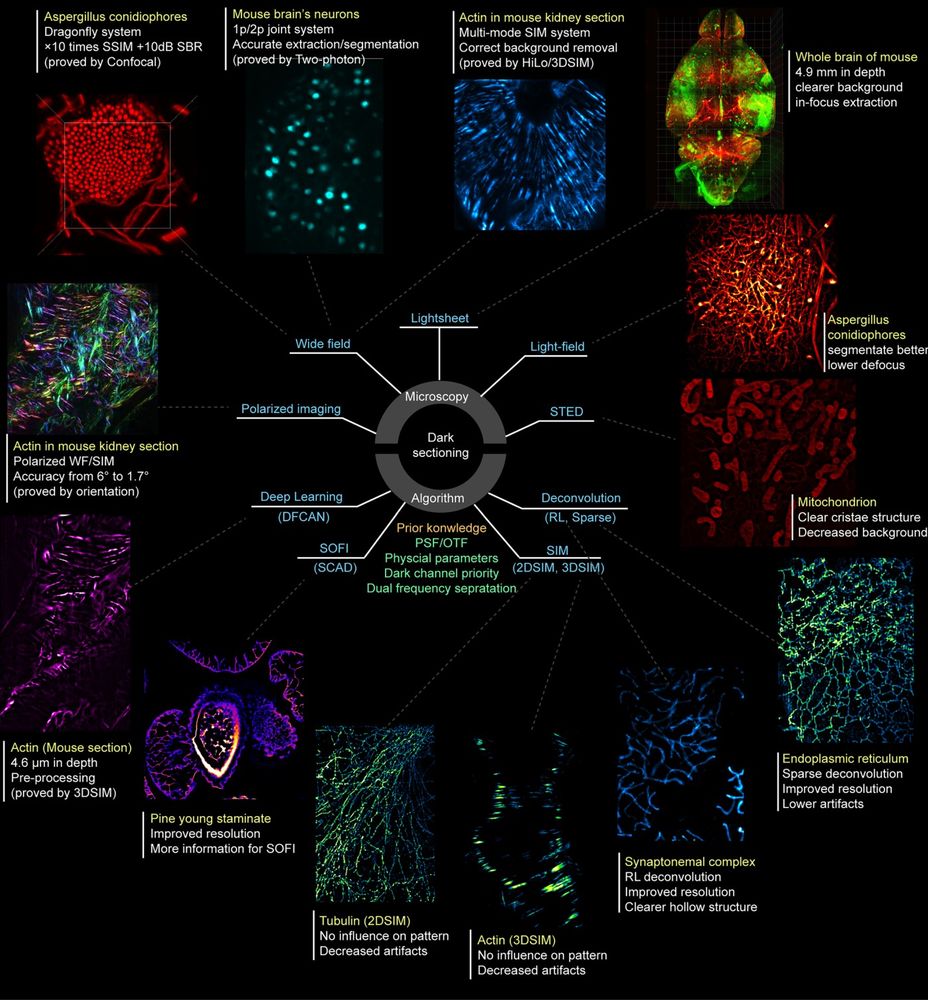
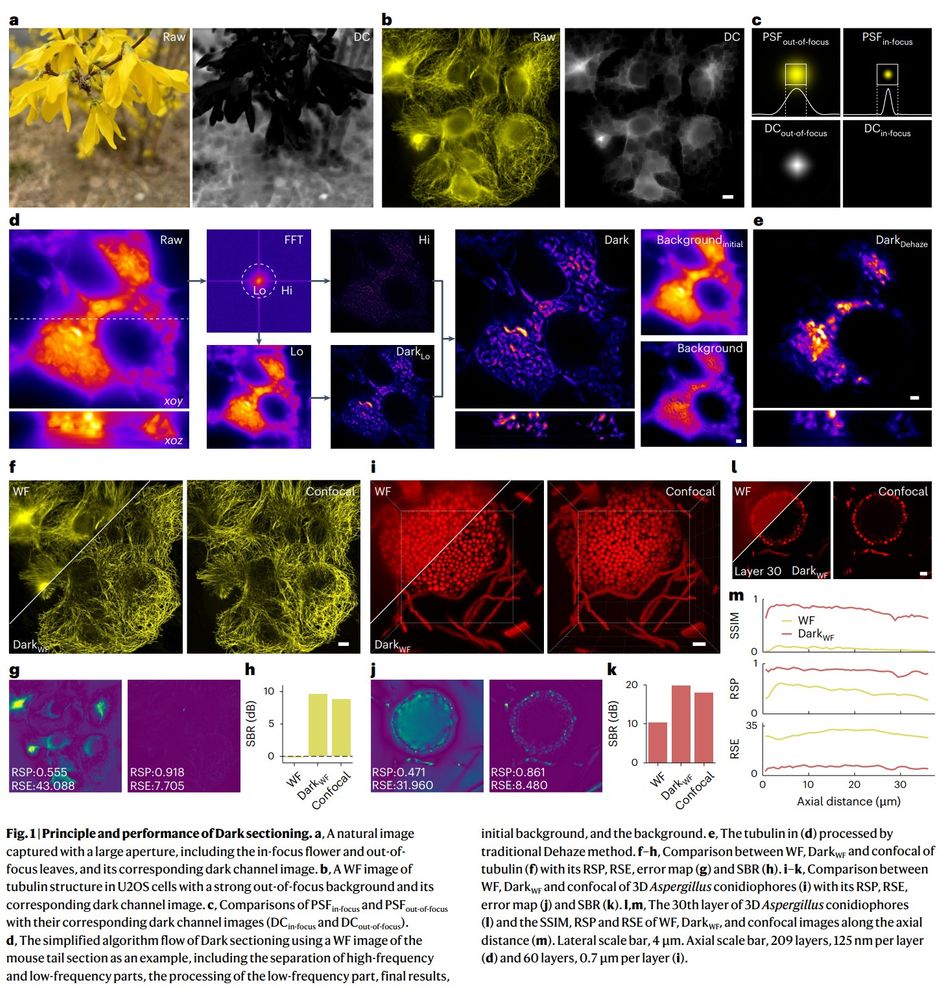
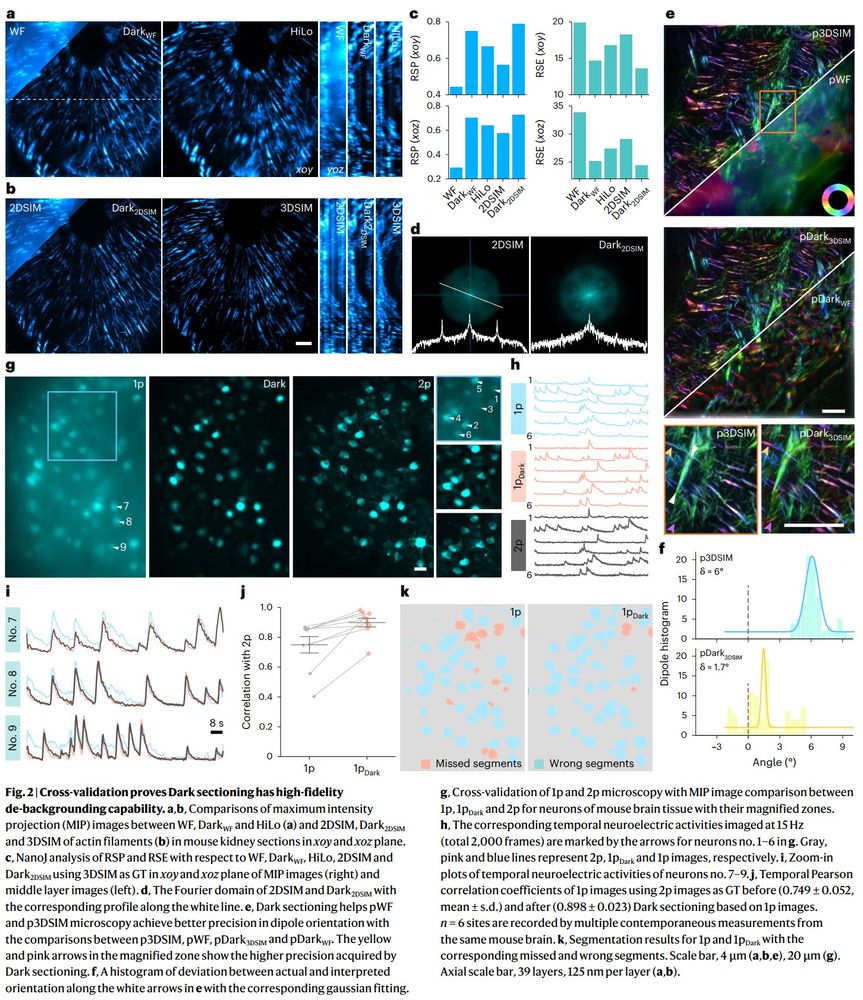
🔬 Widefield → Confocal
🔬 Single-photon → Multiphoton
🔬 2D-SIM → 3D-SIM
Now integrated in Airy PolarSIM and #FlexSIM!
doi.org/10.1038/s415...
#Microscopy #Fluorescence