
Interested in gene regulation in haematopoiesis and AML.

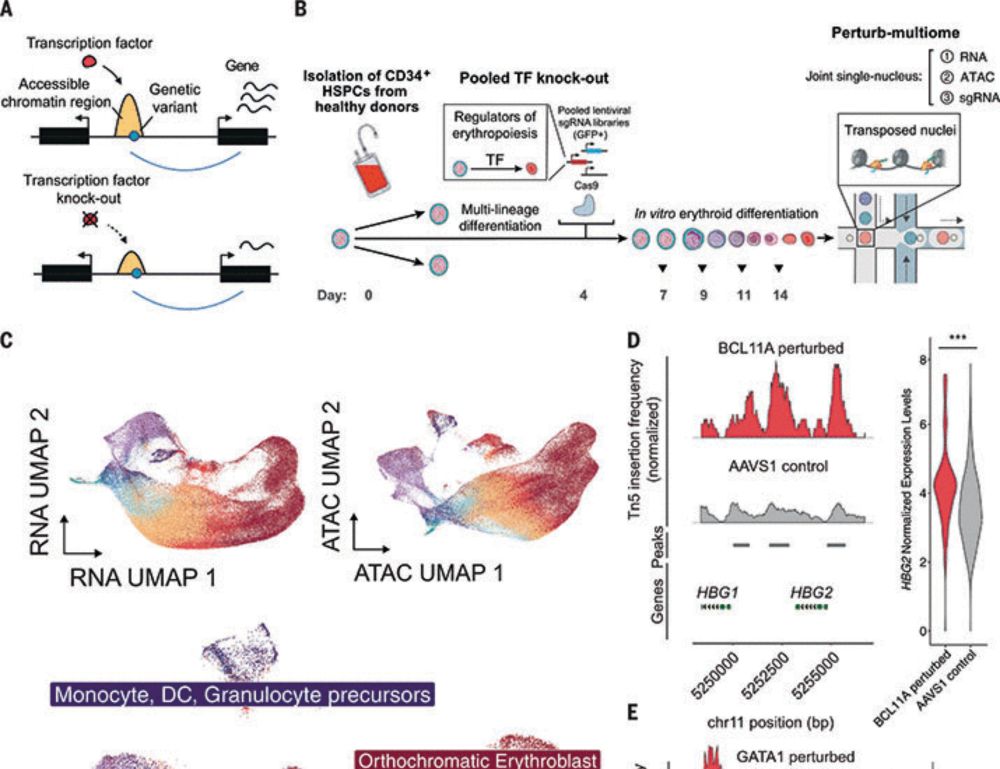
academic.oup.com/nar/article/...
@mrattray.bsky.social @miqbal13.bsky.social @will-morgans.bsky.social

academic.oup.com/nar/article/...
@mrattray.bsky.social @miqbal13.bsky.social @will-morgans.bsky.social




www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
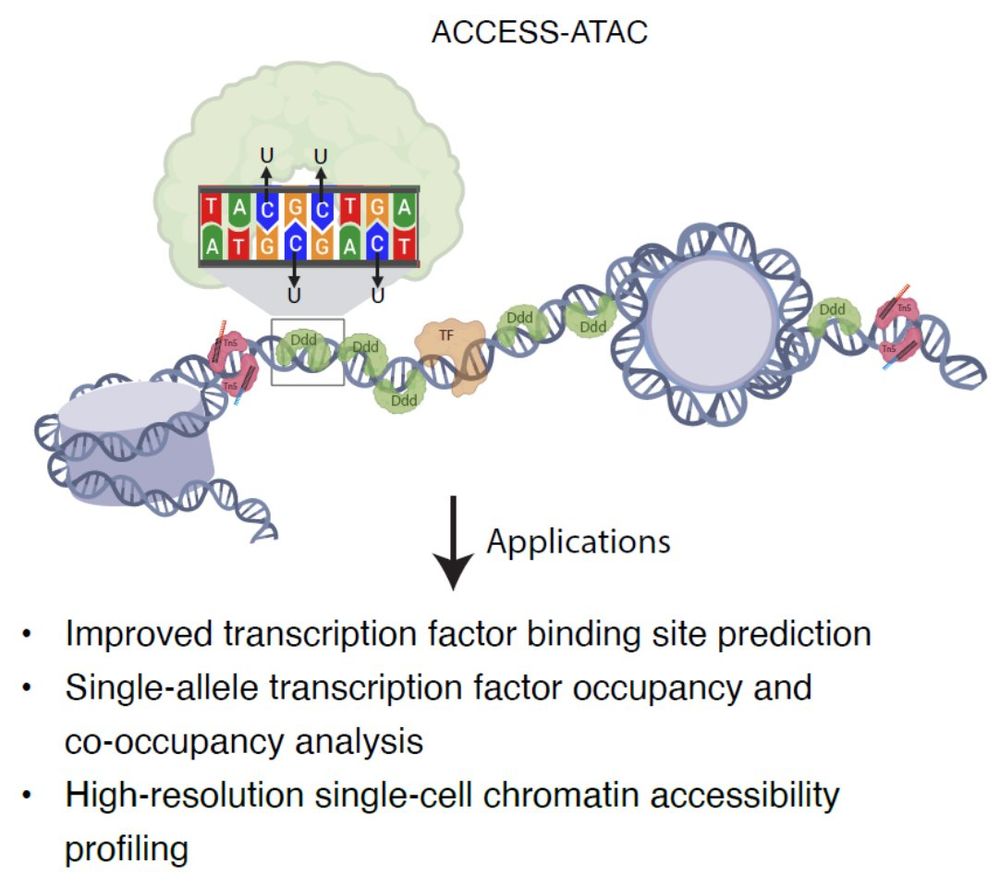
www.biorxiv.org/content/10.1...
biorxiv.org/content/10.1101/2024.12.11.627935v1
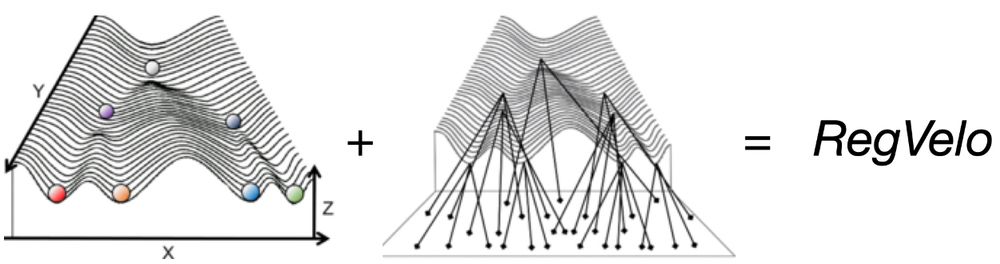
biorxiv.org/content/10.1101/2024.12.11.627935v1
www.nejm.org/doi/full/10....

www.nejm.org/doi/full/10....
www.nature.com/articles/s41...

www.nature.com/articles/s41...

