Shape the future of computational biology 🧵👇
Shape the future of computational biology 🧵👇
www.nature.com/articles/s41...

www.nature.com/articles/s41...


🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
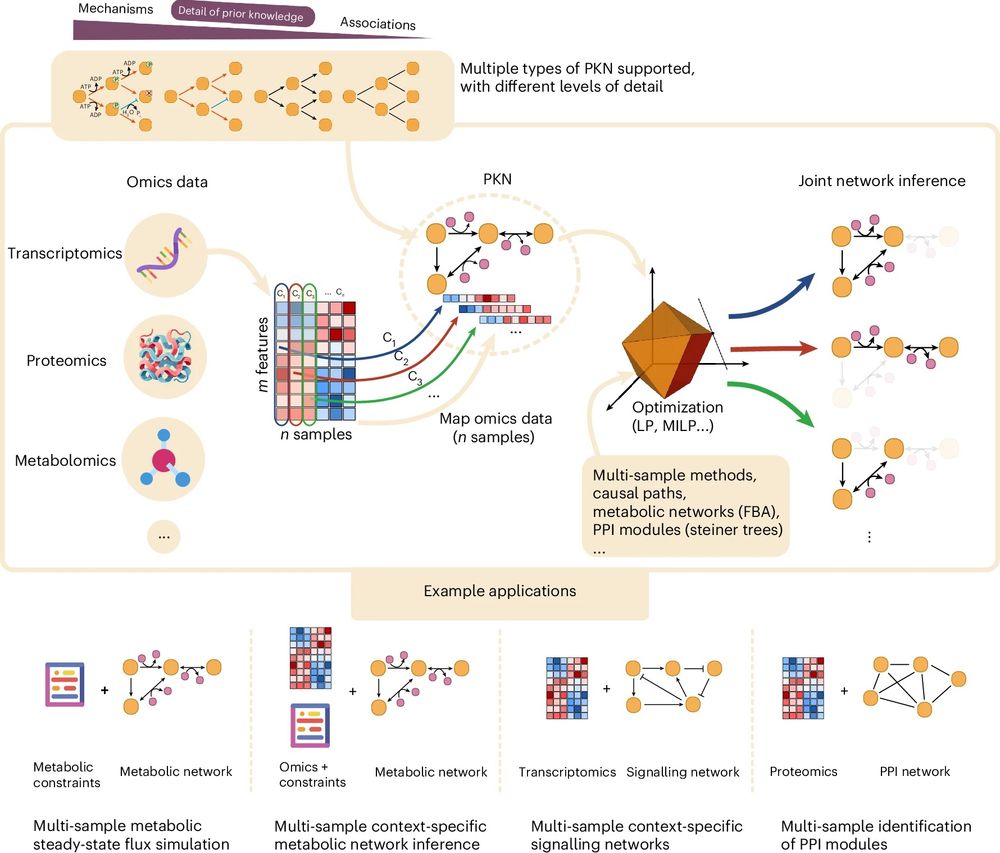
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇

So I built something cool with @anthropic.com's Claude Desktop!
Introducing Claude Text Editor - edit ANY text, ANYWHERE on macOS with Claude's help. No more copy-paste gymnastics! 🚀
So I built something cool with @anthropic.com's Claude Desktop!
Introducing Claude Text Editor - edit ANY text, ANYWHERE on macOS with Claude's help. No more copy-paste gymnastics! 🚀
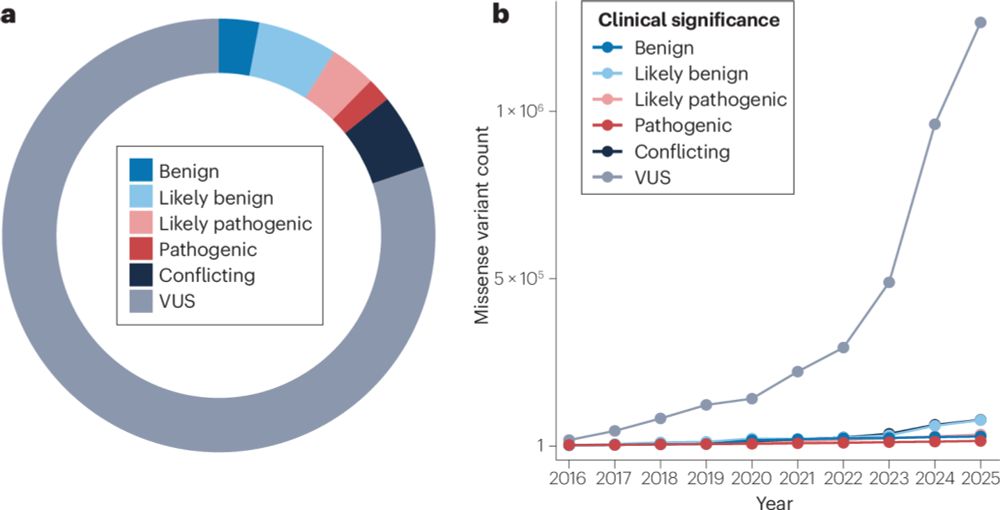
www.nature.com/articles/s41...
By integrating Micro-C with SuperRes Live-Imaging we can calibrate genomics&imaging to perform absolute quantification of looping (e.g. this loop is present 3%)
We quantify mESC 36k loops:
www.biorxiv.org/content/10.1...

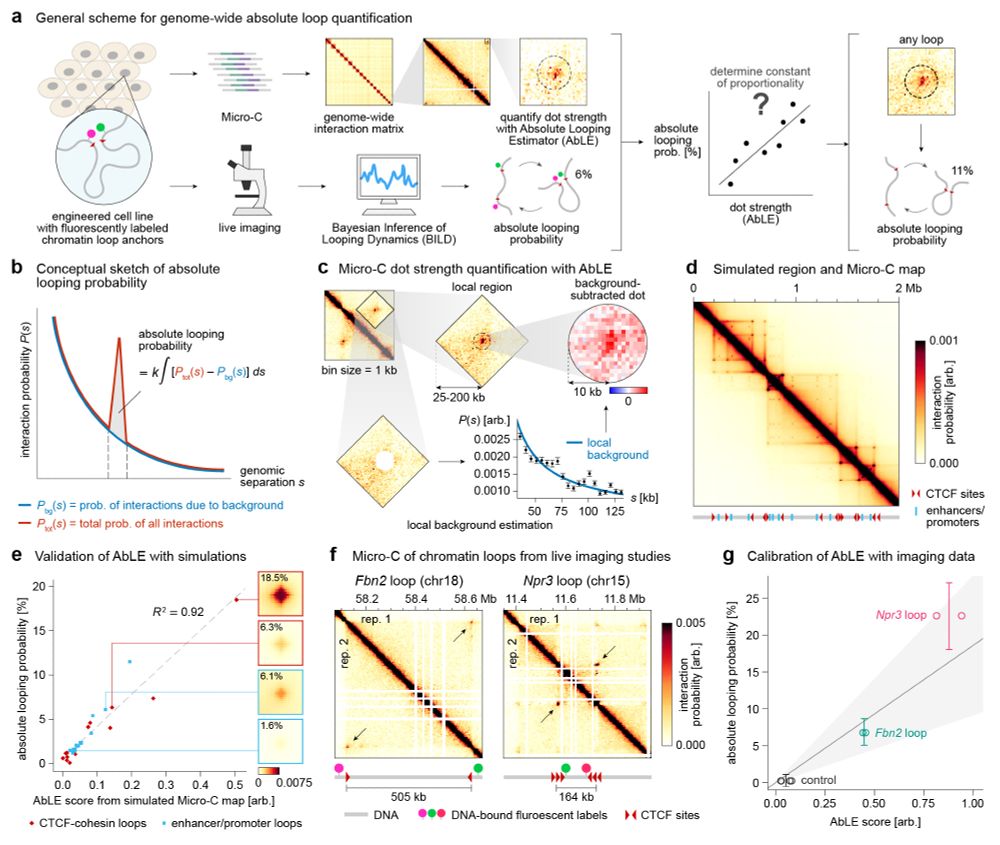
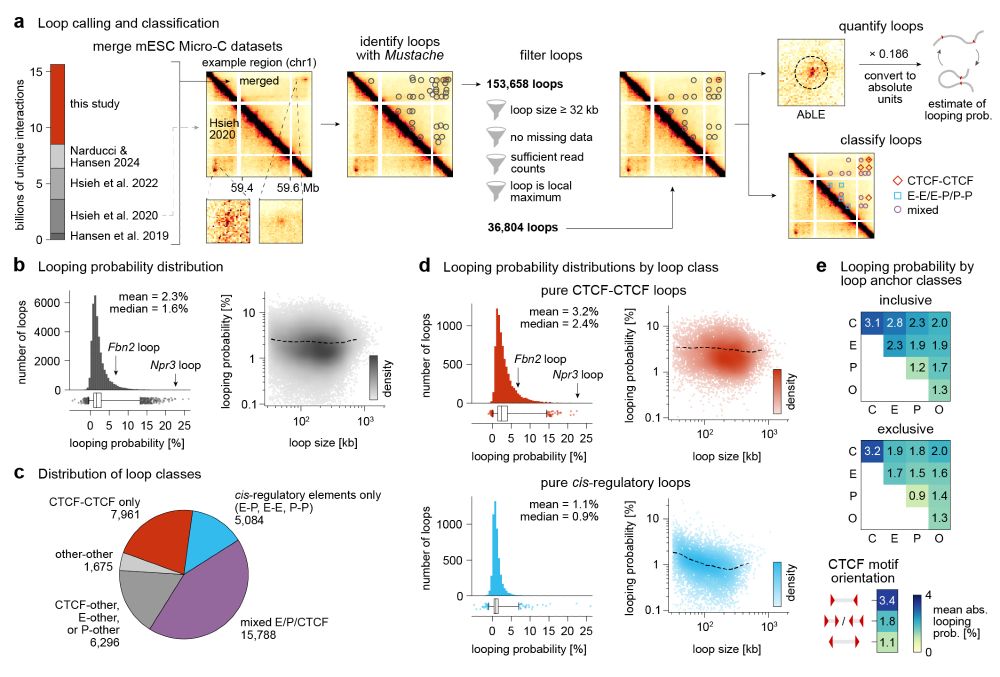
By integrating Micro-C with SuperRes Live-Imaging we can calibrate genomics&imaging to perform absolute quantification of looping (e.g. this loop is present 3%)
We quantify mESC 36k loops:
www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
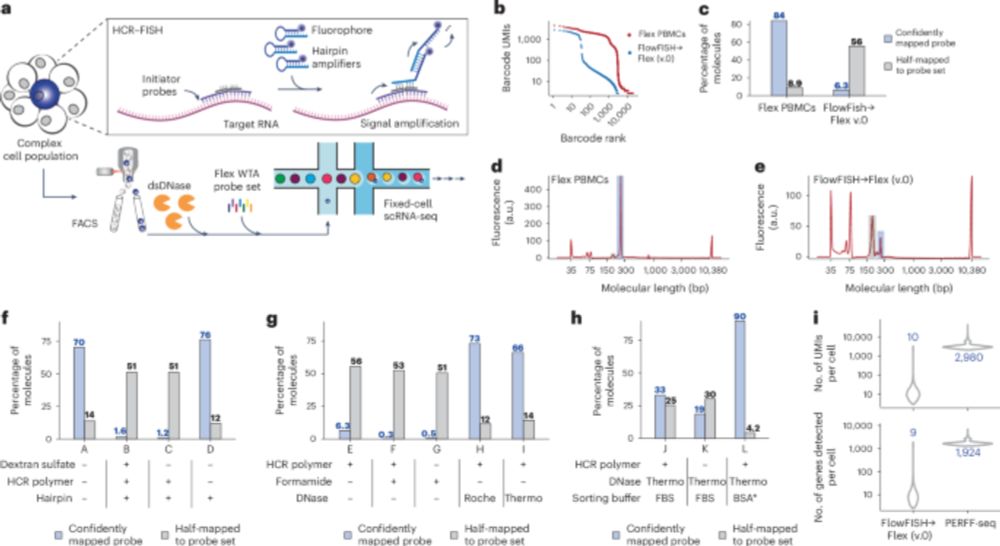
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
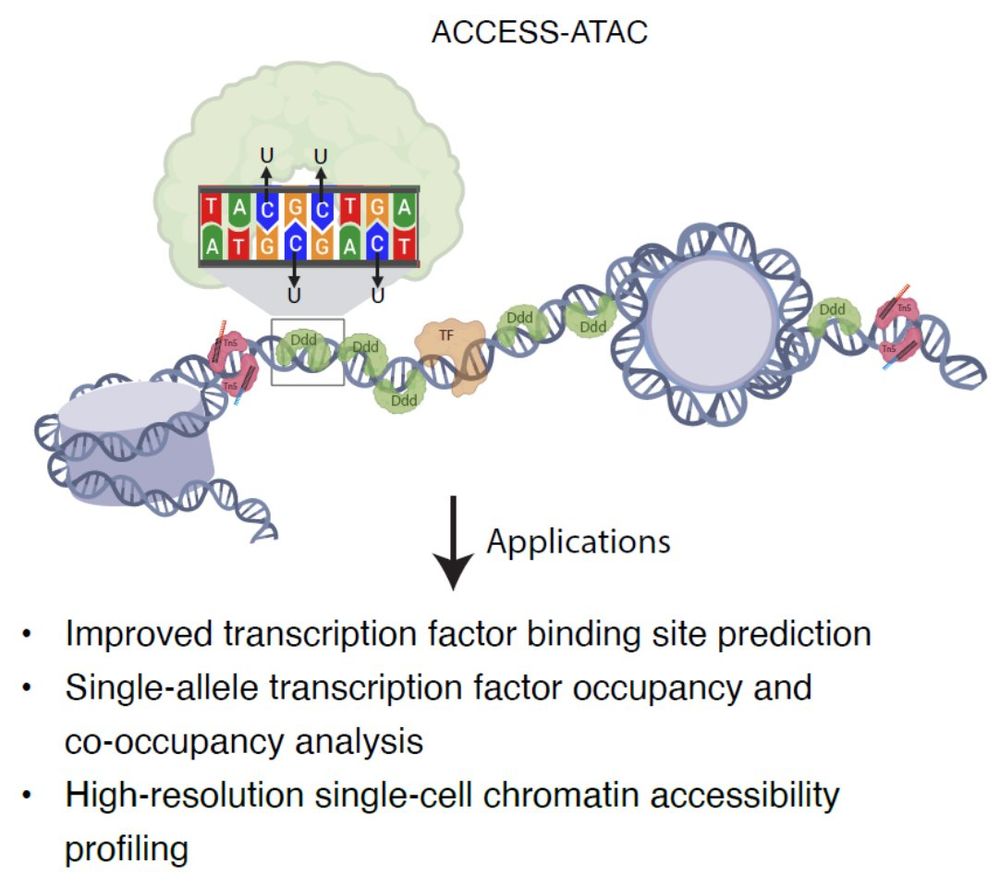
www.biorxiv.org/content/10.1...
@yi-zhang-compbio.bsky.social 🙏
We hope to see you all there 🧬

@yi-zhang-compbio.bsky.social 🙏
www.biorxiv.org/content/10.1...
A 🧵about what it can do:
#SynBio #DeepLearning #GeneRegulation






