Iris Irby
@irisirby.bsky.social
PhD Candidate in the Brown Lab at Georgia Tech | mobile genetic element enthusiast 🧬 | NSF Graduate Research Fellow
Reposted by Iris Irby
🐝🦠 New paper: rdcu.be/eOf7A
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social

Phage diversity mirrors bacterial strain diversity in the honey bee gut microbiota - Nature Communications
Authors analyse paired viral and bacterial shotgun metagenomics data from individual honeybee guts, revealing modular, nested phage–bacteria networks, with viral diversity mirroring bacterial strain c...
www.nature.com
November 4, 2025 at 8:36 PM
🐝🦠 New paper: rdcu.be/eOf7A
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social
Reposted by Iris Irby
New species from space 🦠💫Genomic characterization of Microbacterium meiriae sp. nov., a novel bacterium isolated from the International Space Station www.nature.com/articles/s41...

Genomic characterization of Microbacterium meiriae sp. nov., a novel bacterium isolated from the International Space Station - Scientific Reports
Scientific Reports - Genomic characterization of Microbacterium meiriae sp. nov., a novel bacterium isolated from the International Space Station
www.nature.com
October 18, 2025 at 12:52 PM
New species from space 🦠💫Genomic characterization of Microbacterium meiriae sp. nov., a novel bacterium isolated from the International Space Station www.nature.com/articles/s41...
Reposted by Iris Irby
Phages evolve fast, or do they?
In oysters, some stay identical for years.
With >1,200 phages & 600 Vibrio genomes, we reveal long-term stability and new mobile elements.
Proud of this collaborative work across our teams (Roscoff-UdeM and @epcrocha.bsky.social www.biorxiv.org/cgi/content/...
In oysters, some stay identical for years.
With >1,200 phages & 600 Vibrio genomes, we reveal long-term stability and new mobile elements.
Proud of this collaborative work across our teams (Roscoff-UdeM and @epcrocha.bsky.social www.biorxiv.org/cgi/content/...

Ecological constraints foster both extreme viral-host lineage stability and mobile element diversity in a marine community
Phages are typically viewed as very rapidly evolving biological entities. Little is known, however, about whether and how phages can establish long-term genetic stability. We addressed this eco-evolut...
www.biorxiv.org
October 12, 2025 at 9:16 PM
Phages evolve fast, or do they?
In oysters, some stay identical for years.
With >1,200 phages & 600 Vibrio genomes, we reveal long-term stability and new mobile elements.
Proud of this collaborative work across our teams (Roscoff-UdeM and @epcrocha.bsky.social www.biorxiv.org/cgi/content/...
In oysters, some stay identical for years.
With >1,200 phages & 600 Vibrio genomes, we reveal long-term stability and new mobile elements.
Proud of this collaborative work across our teams (Roscoff-UdeM and @epcrocha.bsky.social www.biorxiv.org/cgi/content/...
Reposted by Iris Irby
New pre-print www.biorxiv.org/content/10.1...
Plasmid-dependent phage (PDPs) are ubiquitous, but the selective pressures that they impose on plasmids are not well understood. Project led by Daniel Cazares in collaboration with @brockhurstlab.bsky.social!
#phagesky#microsky
Plasmid-dependent phage (PDPs) are ubiquitous, but the selective pressures that they impose on plasmids are not well understood. Project led by Daniel Cazares in collaboration with @brockhurstlab.bsky.social!
#phagesky#microsky

Trade-offs between phage resistance and conjugative ability shape the ecological and evolutionary response of a multidrug resistance plasmid to plasmid-dependent phage
Phage therapy is a promising alternative to antibiotics to treat multidrug resistant infections. Plasmid dependent phages (PDPs) are particularly attractive as therapeutics because they can both kill ...
www.biorxiv.org
October 9, 2025 at 10:07 AM
New pre-print www.biorxiv.org/content/10.1...
Plasmid-dependent phage (PDPs) are ubiquitous, but the selective pressures that they impose on plasmids are not well understood. Project led by Daniel Cazares in collaboration with @brockhurstlab.bsky.social!
#phagesky#microsky
Plasmid-dependent phage (PDPs) are ubiquitous, but the selective pressures that they impose on plasmids are not well understood. Project led by Daniel Cazares in collaboration with @brockhurstlab.bsky.social!
#phagesky#microsky
Reposted by Iris Irby
New preprint!
Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...
Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...

Resistance mutation supply modulates the benefit of CRISPR immunity against virulent phages
Only a fraction of bacterial genomes encode CRISPR-Cas systems but the selective causes of this variation are unexplained. How naturally virulent bacteriophages (phages) select for CRISPR immunity has...
www.biorxiv.org
October 6, 2025 at 6:27 AM
New preprint!
Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...
Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...
Reposted by Iris Irby
DYK most P. aeruginosa carry filamentous phage(s) that don't need to kill the cell to reproduce?
We 👉🏻@nanamikubota.bsky.social show that these Pf phages can go ROGUE.
"Filamentous cheater phages drive bacterial and phage populations to lower fitness"
🔗 authors.elsevier.com/c/1lt5I3QW8S...
We 👉🏻@nanamikubota.bsky.social show that these Pf phages can go ROGUE.
"Filamentous cheater phages drive bacterial and phage populations to lower fitness"
🔗 authors.elsevier.com/c/1lt5I3QW8S...

October 2, 2025 at 3:55 PM
DYK most P. aeruginosa carry filamentous phage(s) that don't need to kill the cell to reproduce?
We 👉🏻@nanamikubota.bsky.social show that these Pf phages can go ROGUE.
"Filamentous cheater phages drive bacterial and phage populations to lower fitness"
🔗 authors.elsevier.com/c/1lt5I3QW8S...
We 👉🏻@nanamikubota.bsky.social show that these Pf phages can go ROGUE.
"Filamentous cheater phages drive bacterial and phage populations to lower fitness"
🔗 authors.elsevier.com/c/1lt5I3QW8S...
Reposted by Iris Irby
If you ever find yourself needing evidence for ‘Plasmids are just as common in microbes without resistance genes,’ we’ve got you covered! Check our new paper, out today:
www.microbiologyresearch.org/content/jour...
www.microbiologyresearch.org/content/jour...

Plasmid prevalence is independent of antibiotic resistance in environmental Enterobacteriaceae
The rapid rise of antibiotic-resistant pathogens poses a critical threat to the treatment of infectious diseases. While the spread of antibiotic resistance genes (ARGs) via plasmid conjugation has bee...
www.microbiologyresearch.org
August 12, 2025 at 5:14 PM
If you ever find yourself needing evidence for ‘Plasmids are just as common in microbes without resistance genes,’ we’ve got you covered! Check our new paper, out today:
www.microbiologyresearch.org/content/jour...
www.microbiologyresearch.org/content/jour...
Reposted by Iris Irby
Stoked to finally have a preprint out for Phold, our tool that uses protein structural information to enhance phage genome annotation #phagesky 1/n
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Protein Structure Informed Bacteriophage Genome Annotation with Phold
Bacteriophage (phage) genome annotation is essential for understanding their functional potential and suitability for use as therapeutic agents. Here we introduce Phold, an annotation framework utilis...
www.biorxiv.org
August 8, 2025 at 7:11 AM
Stoked to finally have a preprint out for Phold, our tool that uses protein structural information to enhance phage genome annotation #phagesky 1/n
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Iris Irby
New short story from my postdoc in the Bassler lab is up on bioRxiv!
Turns out our favorite quorum sensing responsive phage VP882 is a member of a larger family of phages that is distributed across multiple bacterial species and can eavesdrop on a universally conserved autoinducer.
#PhageSky 🦠🧫
Turns out our favorite quorum sensing responsive phage VP882 is a member of a larger family of phages that is distributed across multiple bacterial species and can eavesdrop on a universally conserved autoinducer.
#PhageSky 🦠🧫
A family of linear plasmid phages that detect a quorum-sensing autoinducer exists in multiple bacterial species https://www.biorxiv.org/content/10.1101/2025.07.30.667625v1
July 31, 2025 at 10:18 AM
New short story from my postdoc in the Bassler lab is up on bioRxiv!
Turns out our favorite quorum sensing responsive phage VP882 is a member of a larger family of phages that is distributed across multiple bacterial species and can eavesdrop on a universally conserved autoinducer.
#PhageSky 🦠🧫
Turns out our favorite quorum sensing responsive phage VP882 is a member of a larger family of phages that is distributed across multiple bacterial species and can eavesdrop on a universally conserved autoinducer.
#PhageSky 🦠🧫
Reposted by Iris Irby
New pre-print from my lab!
Plasmids link antibiotic resistance and phage defense.
E.coli plasmids are hotspots for both antibiotic resistance and phage defense. Phage therapy has the potential to accidentally select for antibiotic resistance!!!
#microsky#AMR#phage
www.biorxiv.org/content/10.1....
Plasmids link antibiotic resistance and phage defense.
E.coli plasmids are hotspots for both antibiotic resistance and phage defense. Phage therapy has the potential to accidentally select for antibiotic resistance!!!
#microsky#AMR#phage
www.biorxiv.org/content/10.1....

Plasmids link antibiotic resistance genes and phage defense systems in E. coli
Phage therapy has been proposed as an alternative to antibiotics to treat resistant infections. However, we have a limited understanding of how antibiotic resistance genes (ARGs) associate with bacter...
www.biorxiv.org
July 29, 2025 at 1:28 PM
New pre-print from my lab!
Plasmids link antibiotic resistance and phage defense.
E.coli plasmids are hotspots for both antibiotic resistance and phage defense. Phage therapy has the potential to accidentally select for antibiotic resistance!!!
#microsky#AMR#phage
www.biorxiv.org/content/10.1....
Plasmids link antibiotic resistance and phage defense.
E.coli plasmids are hotspots for both antibiotic resistance and phage defense. Phage therapy has the potential to accidentally select for antibiotic resistance!!!
#microsky#AMR#phage
www.biorxiv.org/content/10.1....
Reposted by Iris Irby
After many years in the making, here is our host range #phage paper with #ecology, #evolution and #biocontrol perspectives published in Molecular Ecology! @phimresearch.bsky.social @inrae-pv.bsky.social #PVBMT #JulianGarneau onlinelibrary.wiley.com/doi/10.1111/...
These are our key findings:
These are our key findings:

A Phylogenetic Host‐Range Index Reveals Ecological Constraints in Phage Specialisation and Virulence
Phages are typically known for having a limited host range, targeting particular strains within a bacterial species, but accurately measuring their specificity remains challenging. Factors like the g...
onlinelibrary.wiley.com
July 28, 2025 at 11:13 AM
After many years in the making, here is our host range #phage paper with #ecology, #evolution and #biocontrol perspectives published in Molecular Ecology! @phimresearch.bsky.social @inrae-pv.bsky.social #PVBMT #JulianGarneau onlinelibrary.wiley.com/doi/10.1111/...
These are our key findings:
These are our key findings:
Reposted by Iris Irby
🧪Struggling to induce #prophage?
🧬 Meet PSOSP, a tool to predict prophage induction mode!
🚀Distinguishes SOS-dependent vs independent prophages.
🖥️Available on web server: vee-lab.sjtu.edu.cn/PSOSP and Github: github.com/mujiezhang/P...
#Bioinformatics #phage #viruse #microbiome #GutHealth
🧬 Meet PSOSP, a tool to predict prophage induction mode!
🚀Distinguishes SOS-dependent vs independent prophages.
🖥️Available on web server: vee-lab.sjtu.edu.cn/PSOSP and Github: github.com/mujiezhang/P...
#Bioinformatics #phage #viruse #microbiome #GutHealth
GitHub - mujiezhang/PSOSP: PSOSP (Prophage SOS dependency Predictor) is a novel bioinformatics tool to predict prophage induction modes by analyzing the heterology index (HI) of LexA protein binding t...
PSOSP (Prophage SOS dependency Predictor) is a novel bioinformatics tool to predict prophage induction modes by analyzing the heterology index (HI) of LexA protein binding to target DNA, classifyin...
github.com
July 27, 2025 at 1:05 PM
🧪Struggling to induce #prophage?
🧬 Meet PSOSP, a tool to predict prophage induction mode!
🚀Distinguishes SOS-dependent vs independent prophages.
🖥️Available on web server: vee-lab.sjtu.edu.cn/PSOSP and Github: github.com/mujiezhang/P...
#Bioinformatics #phage #viruse #microbiome #GutHealth
🧬 Meet PSOSP, a tool to predict prophage induction mode!
🚀Distinguishes SOS-dependent vs independent prophages.
🖥️Available on web server: vee-lab.sjtu.edu.cn/PSOSP and Github: github.com/mujiezhang/P...
#Bioinformatics #phage #viruse #microbiome #GutHealth
Reposted by Iris Irby
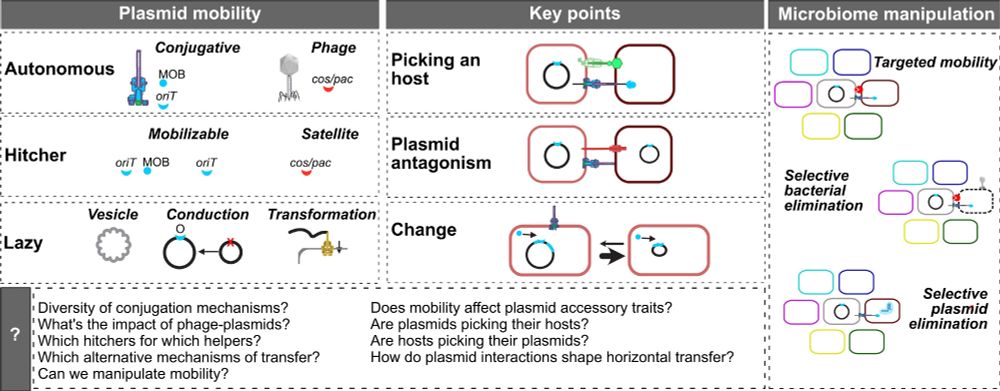
Here's our new broad review on the extended mobility of plasmids, about all mechanisms driving and limiting their transfer. From conjugation to conduction, phage-plasmids to hitchers, molecular to evolutionary dynamics, ecology to biotech. The state of affairs. 1/9 academic.oup.com/nar/article/...
July 23, 2025 at 7:35 AM
Here's our new broad review on the extended mobility of plasmids, about all mechanisms driving and limiting their transfer. From conjugation to conduction, phage-plasmids to hitchers, molecular to evolutionary dynamics, ecology to biotech. The state of affairs. 1/9 academic.oup.com/nar/article/...
Reposted by Iris Irby
Do plasmids evolve faster 🐇, slower 🐢, or just like chromosomes 🧬?
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
July 22, 2025 at 3:05 PM
Do plasmids evolve faster 🐇, slower 🐢, or just like chromosomes 🧬?
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
Reposted by Iris Irby
We made structural predictions of representatives of each #PHROG from #phage genomes, and put the whole lot online. You can browse through montages or go to your favourite phrog directly and download its PDB.
💻🧬 #phagesky
linsalrob.github.io/PHROG_struct...
💻🧬 #phagesky
linsalrob.github.io/PHROG_struct...
PHROG Structure Gallery
linsalrob.github.io
July 7, 2025 at 8:28 AM
We made structural predictions of representatives of each #PHROG from #phage genomes, and put the whole lot online. You can browse through montages or go to your favourite phrog directly and download its PDB.
💻🧬 #phagesky
linsalrob.github.io/PHROG_struct...
💻🧬 #phagesky
linsalrob.github.io/PHROG_struct...
Reposted by Iris Irby
Curious about plasmid biology? Our latest paper is out now in Nature Communications! 🚨
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
Universal rules govern plasmid copy number - Nature Communications
Plasmids exhibit a broad range of sizes and copies per cell, and these two parameters appear to be negatively correlated. Here, Ramiro-Martínez et al. analyse the copy number of thousands of diverse b...
doi.org
July 2, 2025 at 10:07 AM
Curious about plasmid biology? Our latest paper is out now in Nature Communications! 🚨
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
Reposted by Iris Irby
New pre-print out \o/ All about CRISPR, metagenomes, and what you learn when you collect (a lot of) spacers from natural communities, with @apcamargo.bsky.social @urineri.bsky.social @lhug.bsky.social but also Uri Gophna, Nikhil George (not on Bsky I think) & others at JGI doi.org/10.1101/2025...
doi.org
June 13, 2025 at 8:22 PM
New pre-print out \o/ All about CRISPR, metagenomes, and what you learn when you collect (a lot of) spacers from natural communities, with @apcamargo.bsky.social @urineri.bsky.social @lhug.bsky.social but also Uri Gophna, Nikhil George (not on Bsky I think) & others at JGI doi.org/10.1101/2025...
Reposted by Iris Irby
Big day for the Gurney lab, first paper from the lab is now published. @sczerwinski.bsky.social led this work and answered a simple question does phage steering work when other bacteria are around? Tldr: yes! journals.asm.org/doi/epub/10....
Phage steering in the presence of a competing bacterial pathogen
You have to enable JavaScript in your browser's settings in order to use the eReader.
journals.asm.org
June 10, 2025 at 5:22 PM
Big day for the Gurney lab, first paper from the lab is now published. @sczerwinski.bsky.social led this work and answered a simple question does phage steering work when other bacteria are around? Tldr: yes! journals.asm.org/doi/epub/10....
Reposted by Iris Irby
Our new big review on Pseudomonas aeruginosa. Hope you enjoy reading it. www.nature.com/articles/s41...
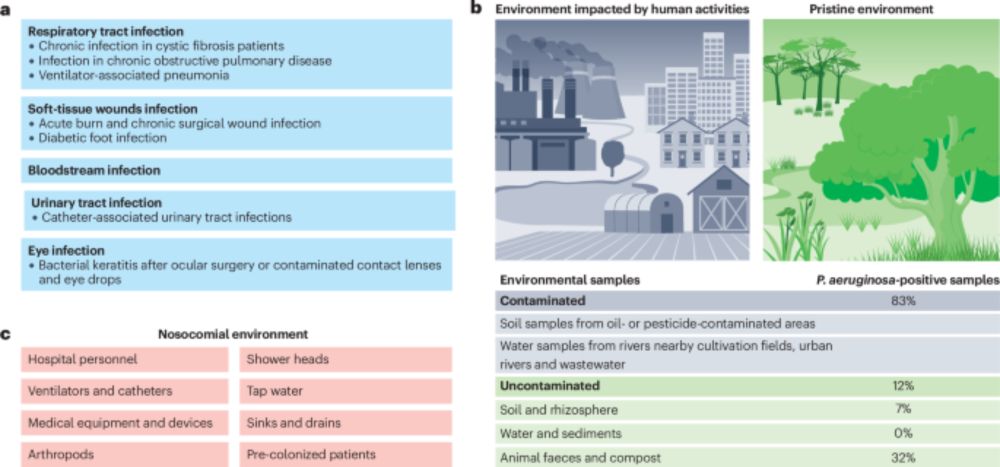
Pseudomonas aeruginosa: ecology, evolution, pathogenesis and antimicrobial susceptibility - Nature Reviews Microbiology
Pseudomonas aeruginosa is a functionally versatile bacterium, a leading opportunistic human pathogen and a model organism in microbiology. In this Review, Letizia, Diggle and Whiteley discuss P. aerug...
www.nature.com
May 29, 2025 at 1:18 PM
Our new big review on Pseudomonas aeruginosa. Hope you enjoy reading it. www.nature.com/articles/s41...
Reposted by Iris Irby
Through the new PLASFIGHTER project, we drive significant advances in unravelling and fighting AMR.
We study plasmid-bacteria interactions to answer important questions about their role in generating and maintaining superbugs.
Discover the project ➡️ plasmidlab.es/project/
#AntibioticResistance
We study plasmid-bacteria interactions to answer important questions about their role in generating and maintaining superbugs.
Discover the project ➡️ plasmidlab.es/project/
#AntibioticResistance

May 28, 2025 at 9:47 AM
Through the new PLASFIGHTER project, we drive significant advances in unravelling and fighting AMR.
We study plasmid-bacteria interactions to answer important questions about their role in generating and maintaining superbugs.
Discover the project ➡️ plasmidlab.es/project/
#AntibioticResistance
We study plasmid-bacteria interactions to answer important questions about their role in generating and maintaining superbugs.
Discover the project ➡️ plasmidlab.es/project/
#AntibioticResistance
Reposted by Iris Irby
Excited to see this new, and extensively-rewritten, version of our preprint up now! With @paulturnerlab.bsky.social we show how bacterial population dynamics (e.g. growth curves) can be used to accurately quantify bacteriophage infectivity
🦠 🧫🔬 #microbiology #VirEvol
doi.org/10.1101/2023...
🦠 🧫🔬 #microbiology #VirEvol
doi.org/10.1101/2023...

Quantifying phage infectivity from characteristics of bacterial population dynamics
A frequent goal of phage biology is to quantify how well a phage kills a population of host bacteria. Unfortunately, traditional methods to quantify phage success can be time-consuming, limiting the t...
doi.org
May 27, 2025 at 10:41 PM
Excited to see this new, and extensively-rewritten, version of our preprint up now! With @paulturnerlab.bsky.social we show how bacterial population dynamics (e.g. growth curves) can be used to accurately quantify bacteriophage infectivity
🦠 🧫🔬 #microbiology #VirEvol
doi.org/10.1101/2023...
🦠 🧫🔬 #microbiology #VirEvol
doi.org/10.1101/2023...
Reposted by Iris Irby
Defence Systems are so hot right now 🔥 🔥 🔥 so check out our latest preprint to see how DSes contribute to niche adaptation, interact with each other, and drive accessory genome interactions in Pseudomonas aeruginosa 1/n
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Defence systems drive accessory genome interactions in Pseudomonas aeruginosa
As well as undergoing mutational selection, bacterial genomes are shaped by a complex evolutionary interplay among diverse accessory genome elements (AGEs). In this study we define AGEs as encompassin...
www.biorxiv.org
May 23, 2025 at 6:33 PM
Defence Systems are so hot right now 🔥 🔥 🔥 so check out our latest preprint to see how DSes contribute to niche adaptation, interact with each other, and drive accessory genome interactions in Pseudomonas aeruginosa 1/n
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Iris Irby
Preprint: Conjugative transfer inhibition of IncA and IncC plasmids by pervasive SGI1-like elements via relaxosome assembly interference
#plasmid #relaxase #bacteria #parasite #MGE #plasmidbiology
www.biorxiv.org/content/10.1...
#plasmid #relaxase #bacteria #parasite #MGE #plasmidbiology
www.biorxiv.org/content/10.1...

Conjugative transfer inhibition of IncA and IncC plasmids by pervasive SGI1-like elements via relaxosome assembly interference
Broad-host-range IncA and IncC conjugative plasmids propagate multidrug resistance in bacteria and mobilize chromosomal resistance islands, including Salmonella Genomic Island 1 (SGI1) across genera. ...
www.biorxiv.org
April 23, 2025 at 3:24 PM
Preprint: Conjugative transfer inhibition of IncA and IncC plasmids by pervasive SGI1-like elements via relaxosome assembly interference
#plasmid #relaxase #bacteria #parasite #MGE #plasmidbiology
www.biorxiv.org/content/10.1...
#plasmid #relaxase #bacteria #parasite #MGE #plasmidbiology
www.biorxiv.org/content/10.1...
Reposted by Iris Irby
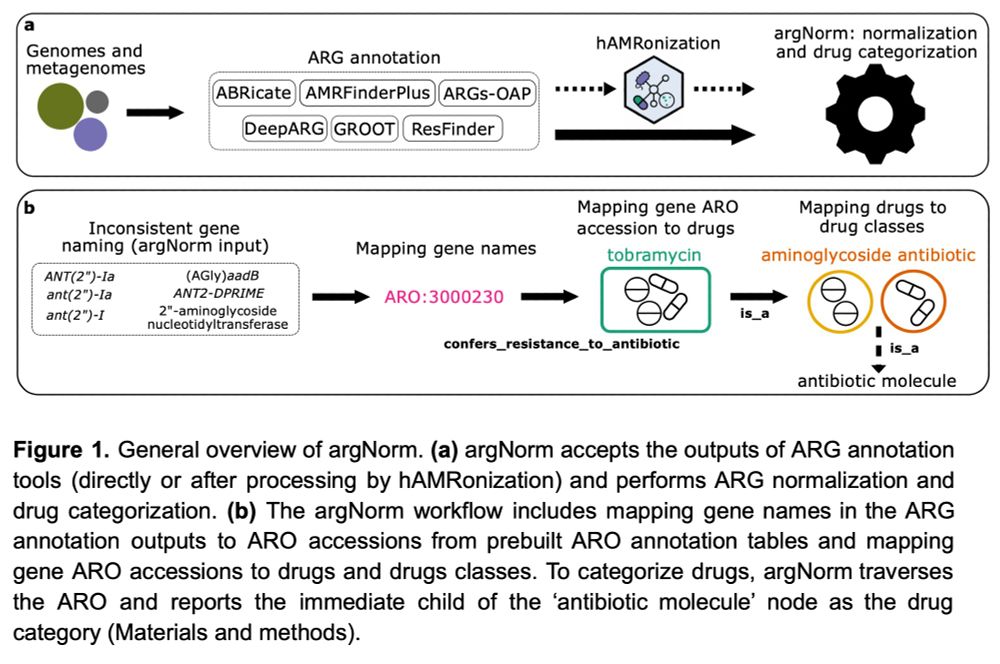
Hey #AMR people,
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6
October 10, 2024 at 7:56 AM
Hey #AMR people,
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6
Reposted by Iris Irby
New paper @isme-microbes.bsky.social : Among a sample of 1044 vibrio phages, we found 17 new Schizotequatrovirus with large genomes (>250kb), a broad host range, and yet a low frequency in our samples (?!). #MicroSky #PhageSky
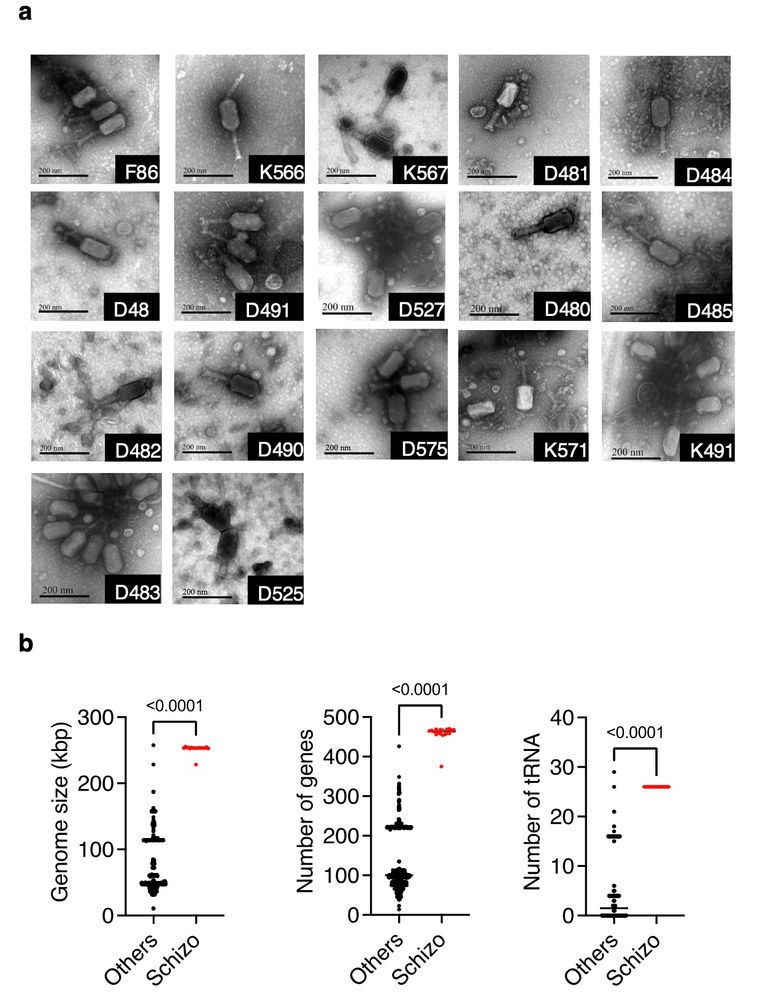
April 14, 2025 at 8:36 AM
New paper @isme-microbes.bsky.social : Among a sample of 1044 vibrio phages, we found 17 new Schizotequatrovirus with large genomes (>250kb), a broad host range, and yet a low frequency in our samples (?!). #MicroSky #PhageSky

